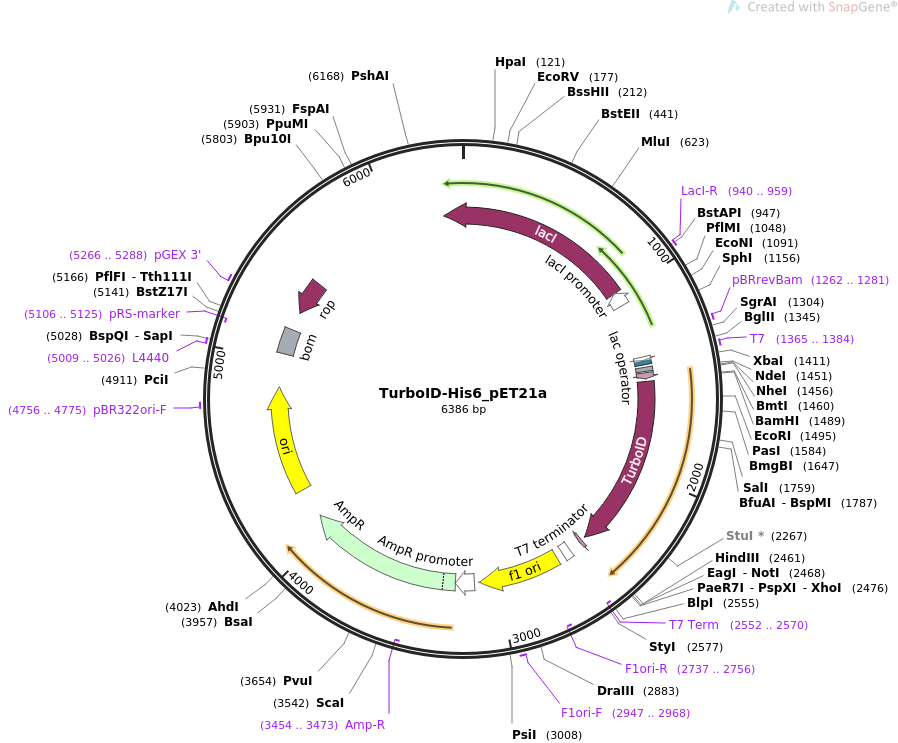
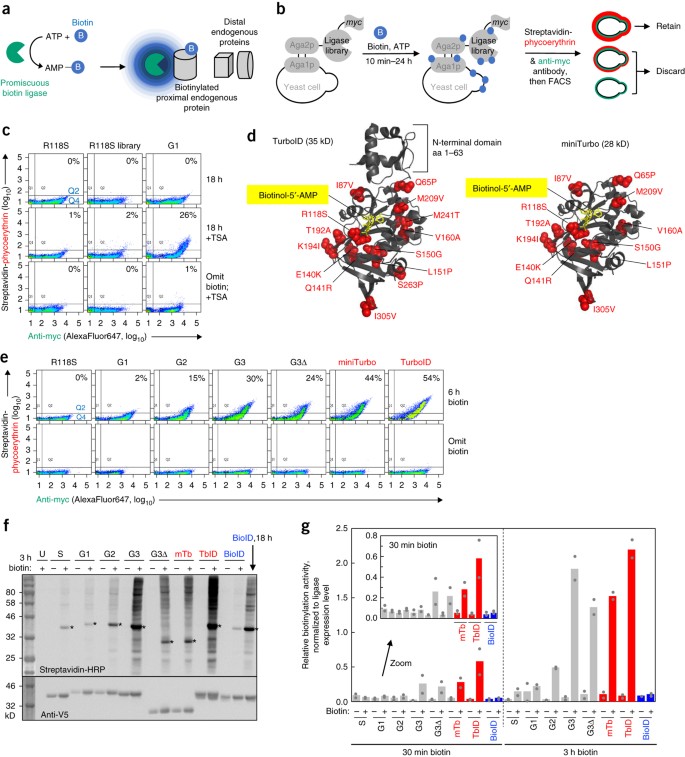
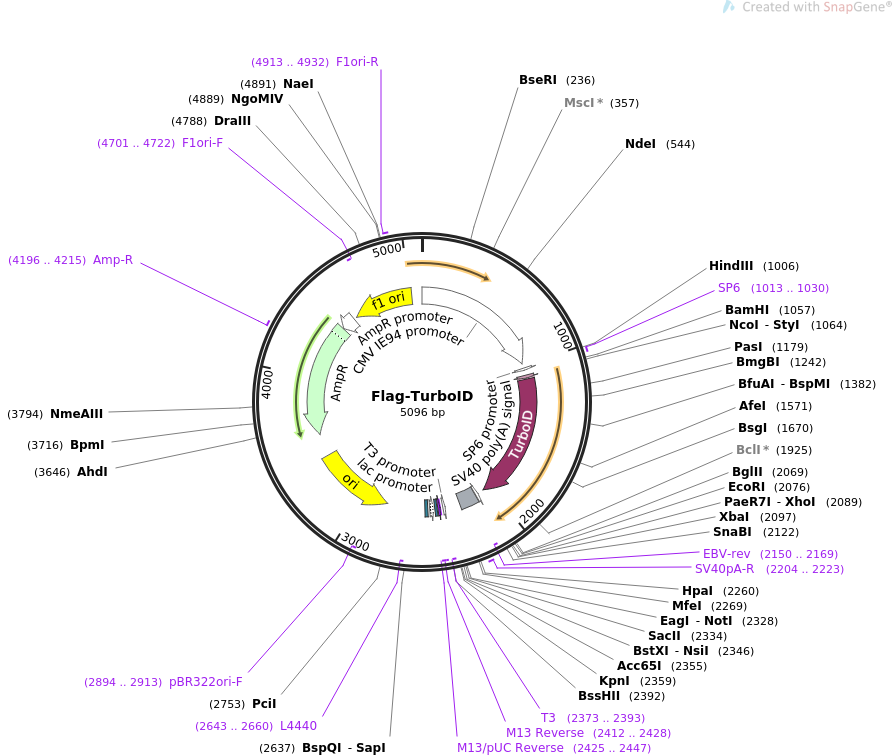
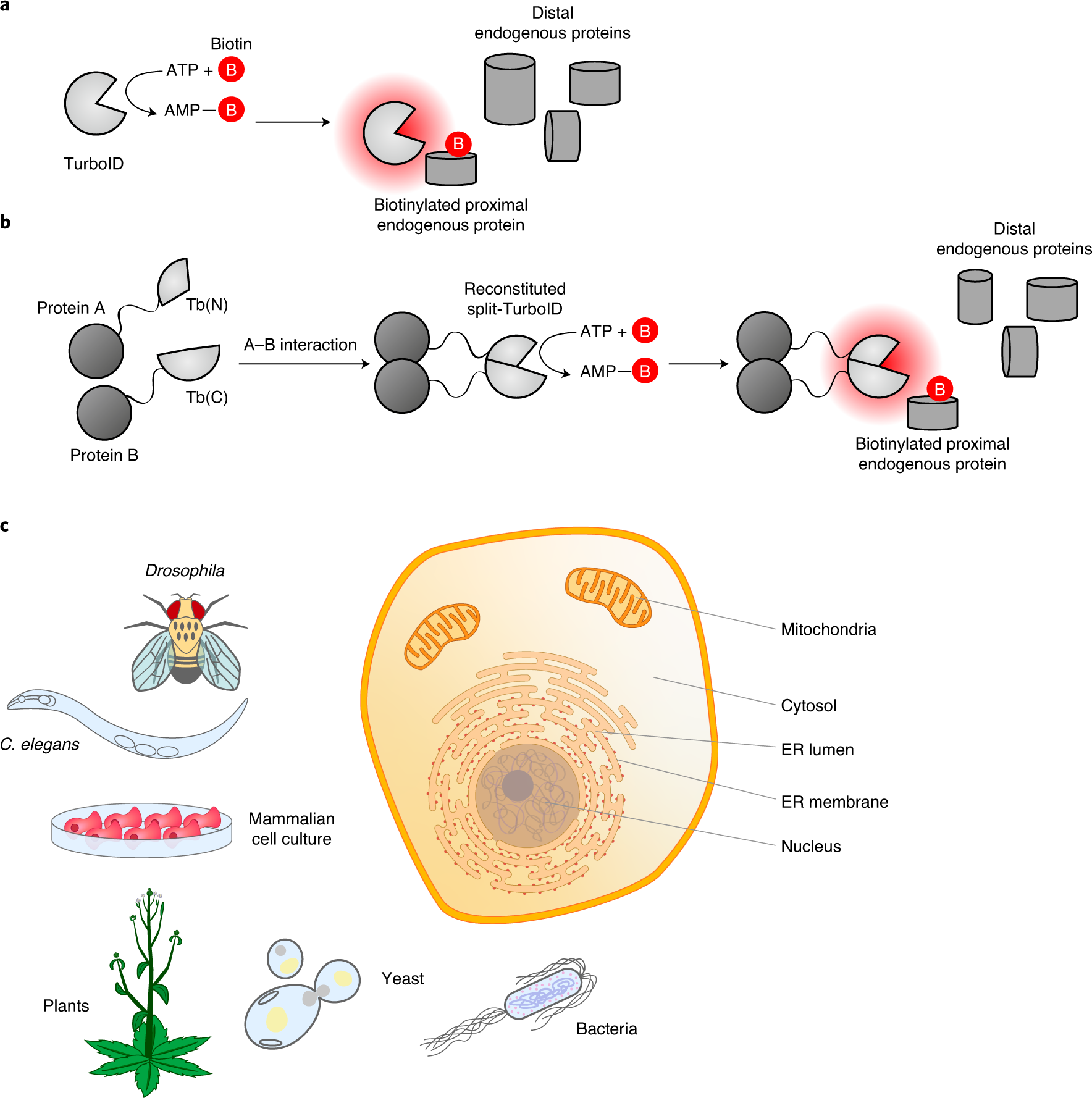
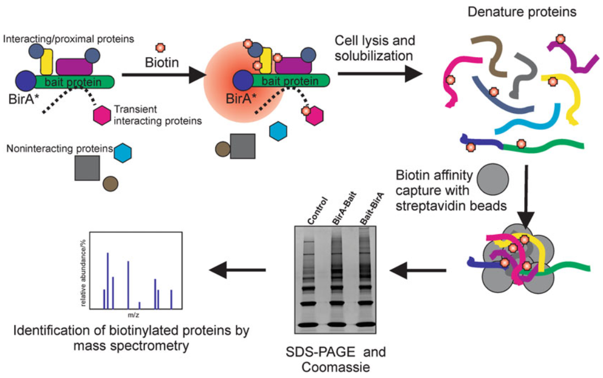
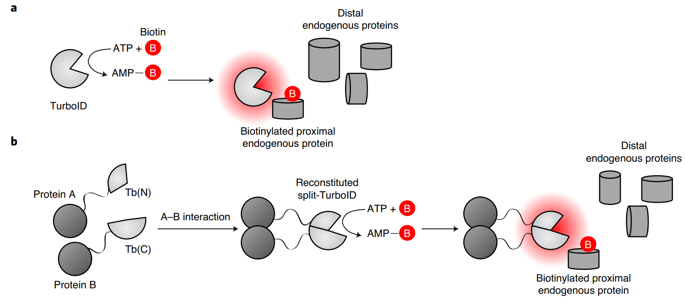
Directed evolution of TurboID for efficient proximity labeling in living cells and organisms | bioRxiv
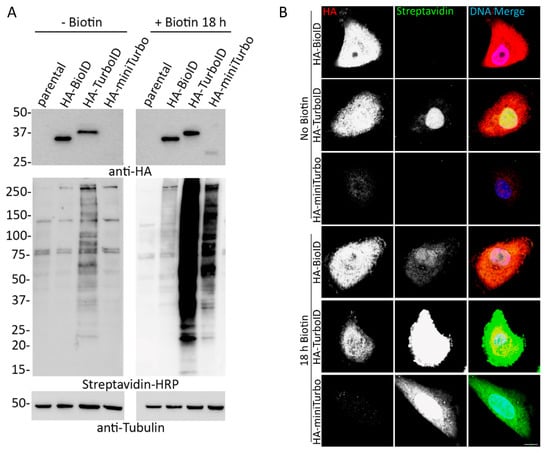
Cells | Free Full-Text | Comparative Application of BioID and TurboID for Protein-Proximity Biotinylation
Establishment of in vivo proximity labeling with biotin using TurboID in the lamentous fungus Sordaria macrospora

Identification of proteomic landscape of drug-binding proteins in live cells by proximity-dependent target ID - ScienceDirect

TurboID-Based Proximity Labeling for In Planta Identification of Protein- Protein Interaction Networks. - Abstract - Europe PMC
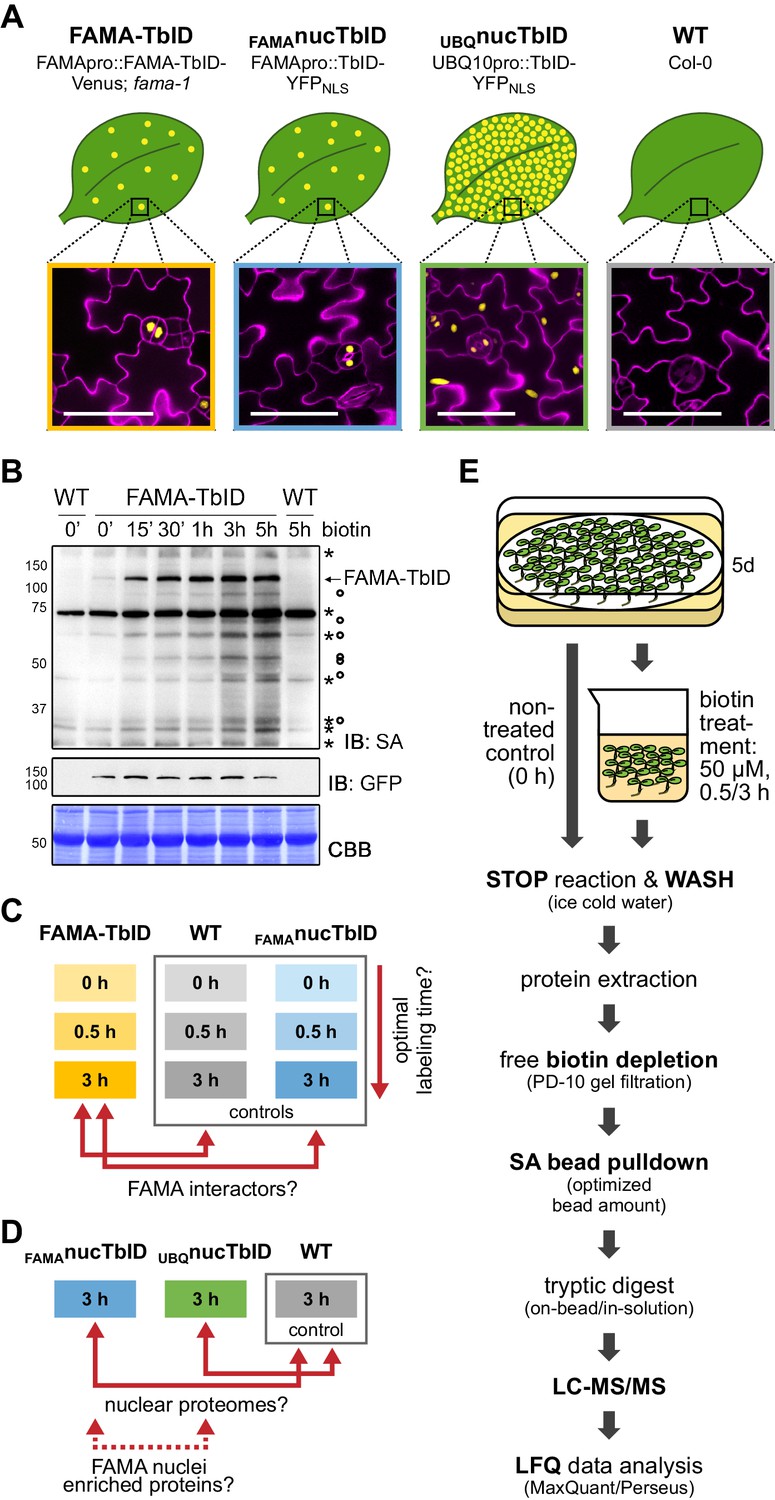
Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID | eLife
Proximity-dependent biotinylation mediated by TurboID to identify protein– protein interaction networks in yeast

TurboID Screening of the OmpP2 Protein Reveals Host Proteins Involved in Recognition and Phagocytosis of Glaesserella parasuis by iPAM Cells | Microbiology Spectrum

Application of TurboID-mediated proximity labeling for mapping a GSK3 kinase signaling network in Arabidopsis | bioRxiv