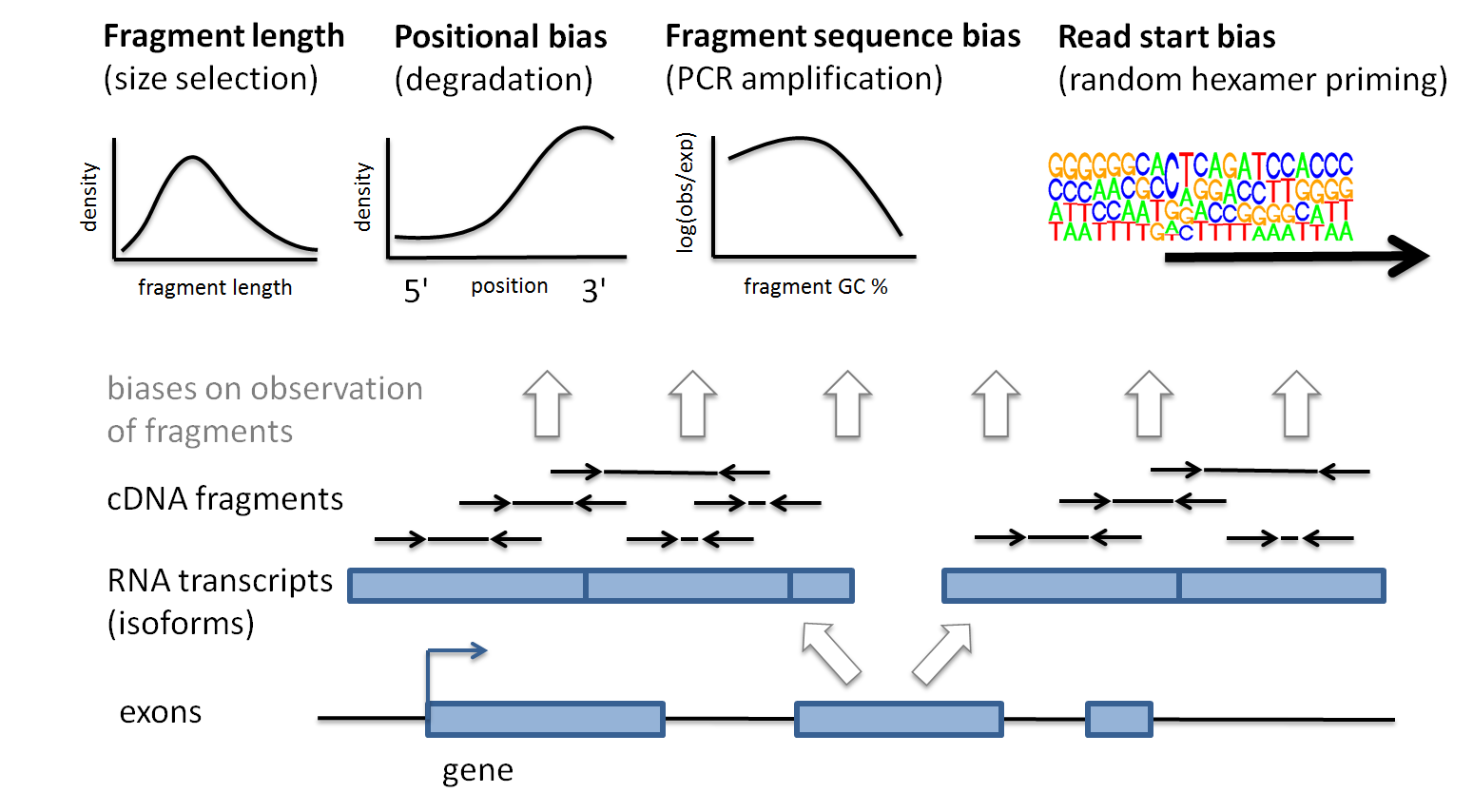
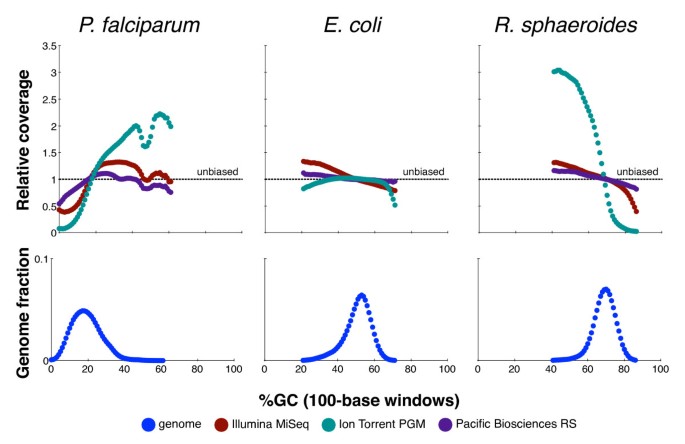
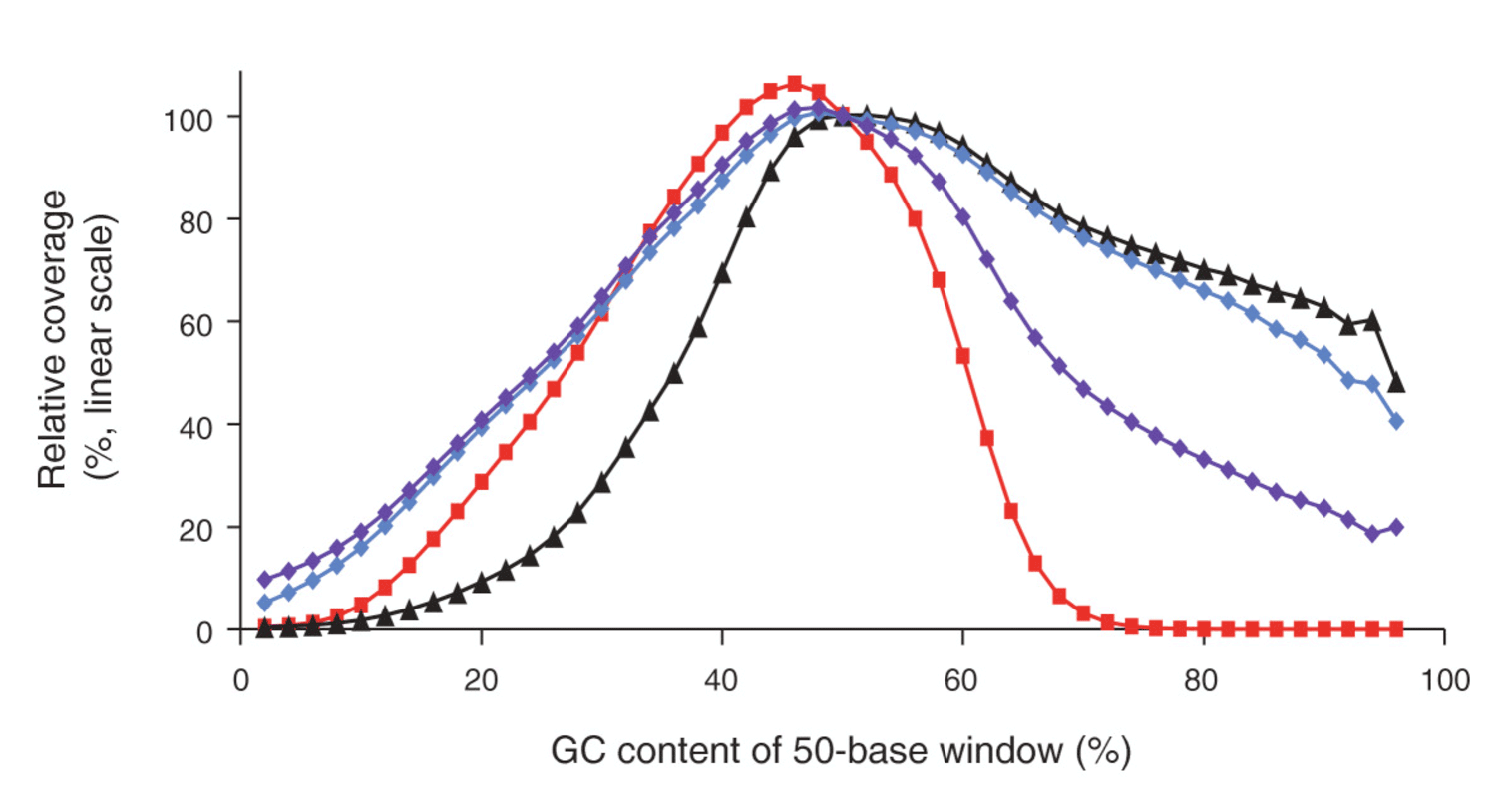
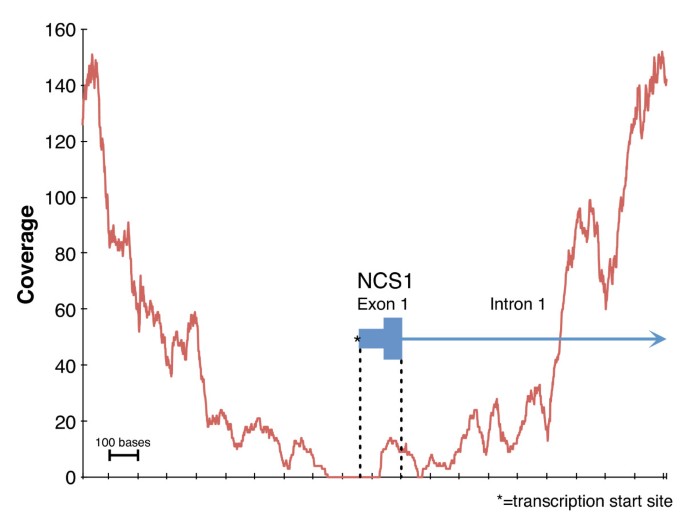
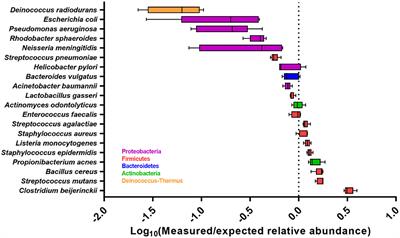
Measuring Illumina Size Bias Using REcount: A Novel Method for Highly Accurate Quantification of Engineered Genetic Constructs | bioRxiv

GC Bias. (A) Schematic of two cycles of GC biased PCR. A sequence with... | Download Scientific Diagram
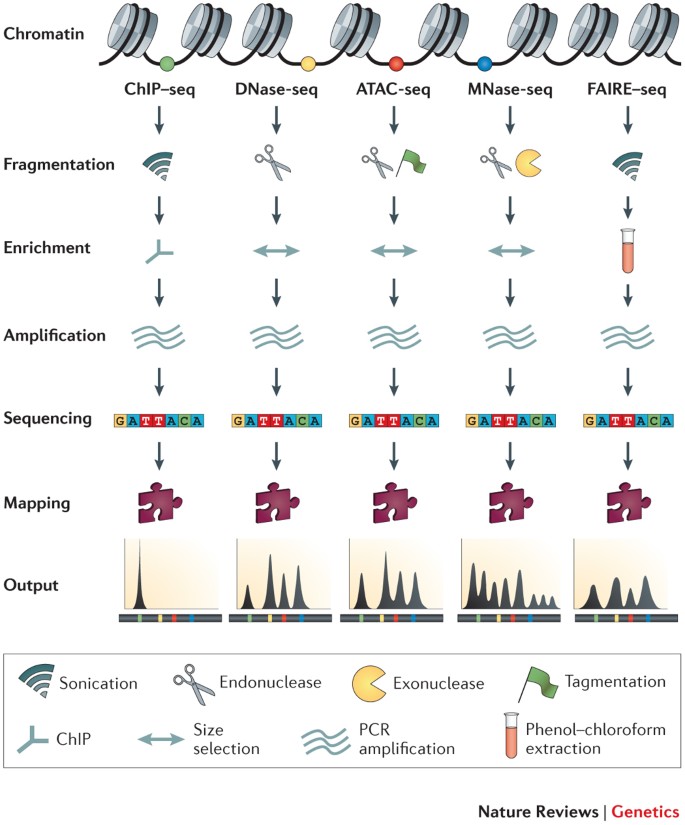
Identifying and mitigating bias in next-generation sequencing methods for chromatin biology | Nature Reviews Genetics

Sensitive and Low-Bias Transcriptome Sequencing Using Agarose PCR | ACS Applied Materials & Interfaces

Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes | PNAS
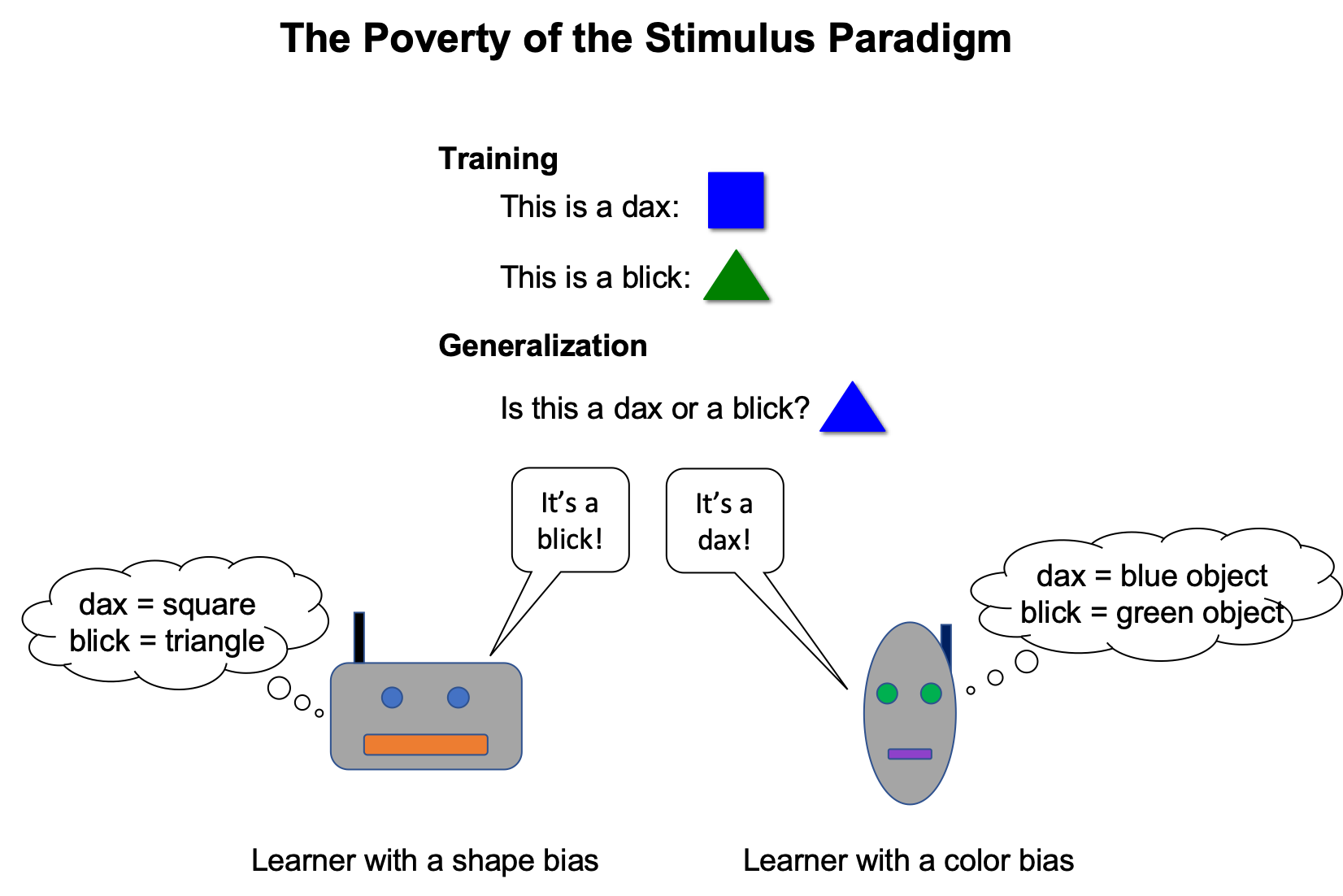
Does Syntax Need to Grow on Trees? Sources of Hierarchical Inductive Bias in Sequence-to-Sequence Networks

Modeling Enzyme Processivity Reveals that RNA-Seq Libraries Are Biased in Characteristic and Correctable Ways: Cell Systems
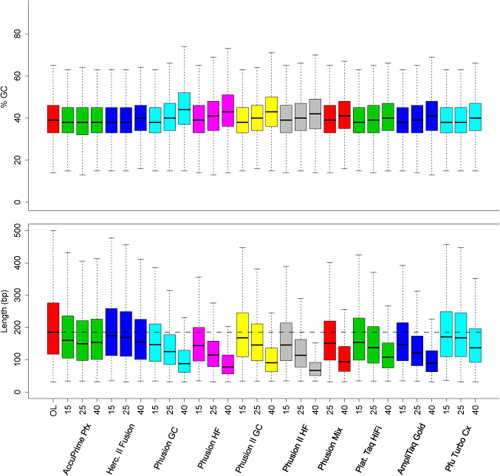
Length and GC-biases during sequencing library amplification: A comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries | BioTechniques