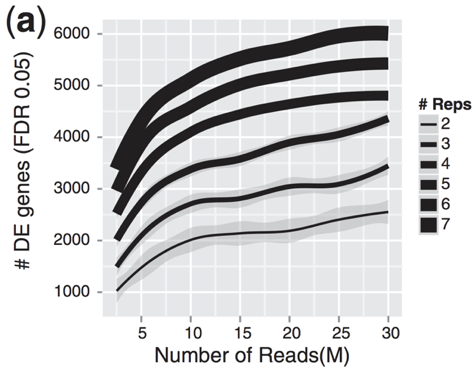
Experimental design considerations | Introduction to RNA-Seq using high-performance computing - ARCHIVED

Recommendations for accurate genotyping of SARS-CoV-2 using amplicon-based sequencing of clinical samples - ScienceDirect
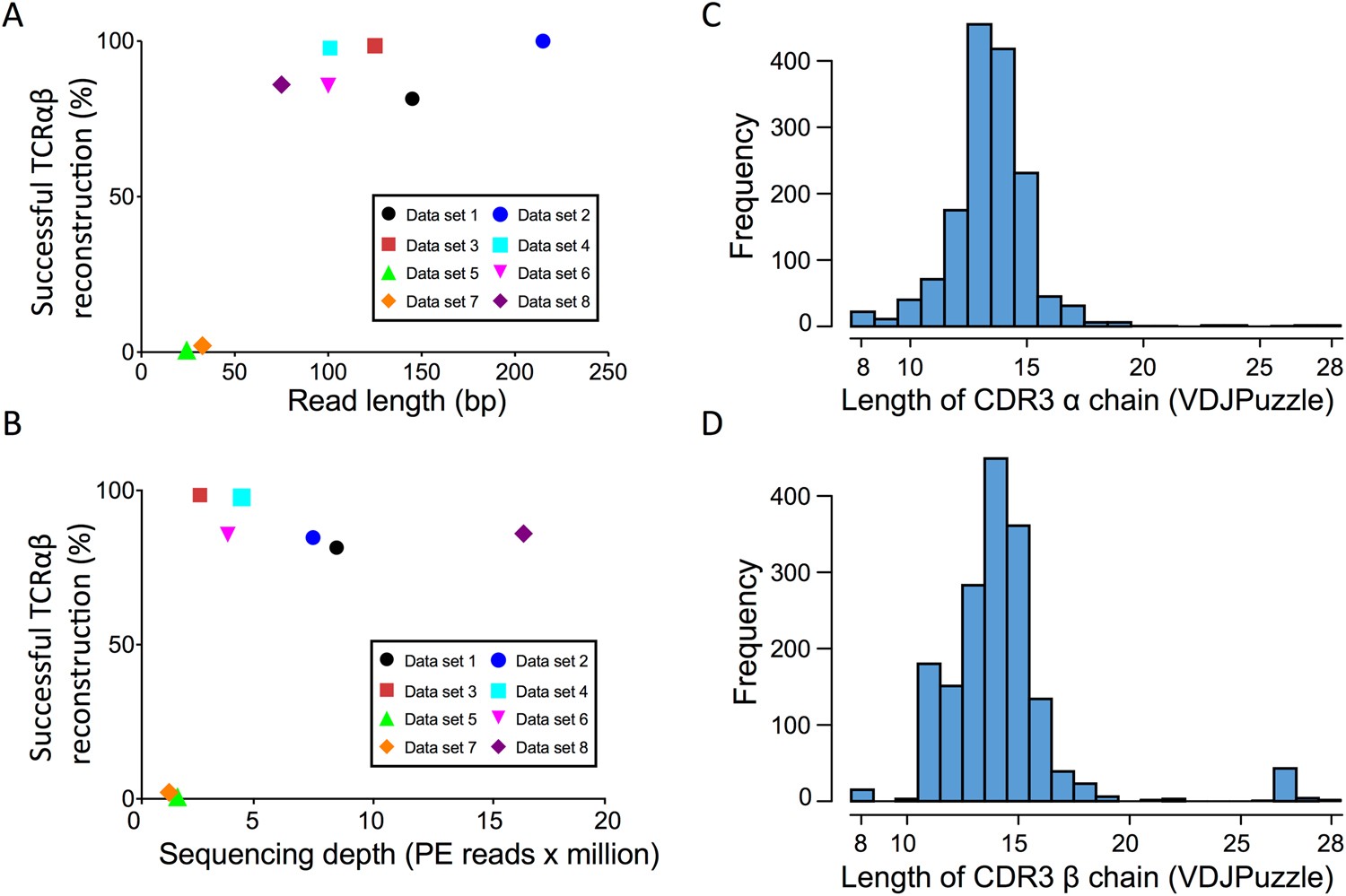
Impact of sequencing depth and read length on single cell RNA sequencing data of T cells | Scientific Reports

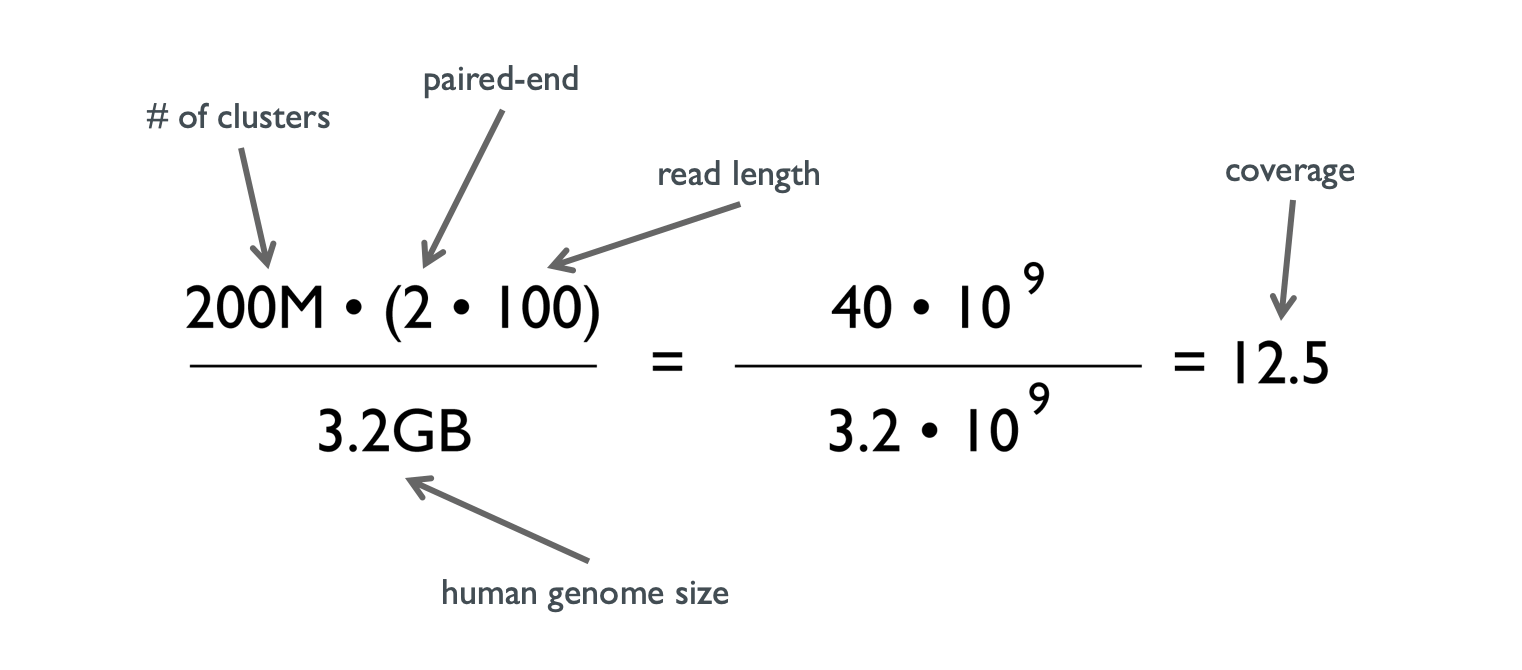
Difference between sequencing Coverage and depth. Depth vs Coverage. Why they are important? - YouTube

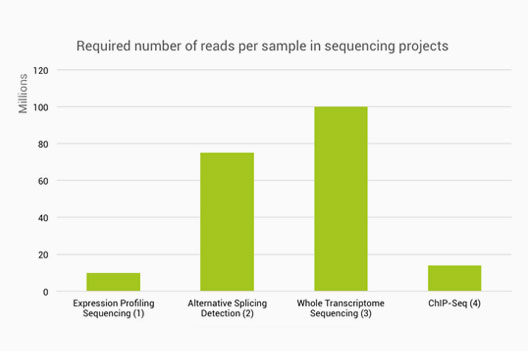
ScienceVision - Different RNA-Seq experiment types require different sequencing read lengths and depth (number of reads per sample). This bulletin reviews RNA sequencing considerations and offers resources for planning RNA-Seq experiments. Link:




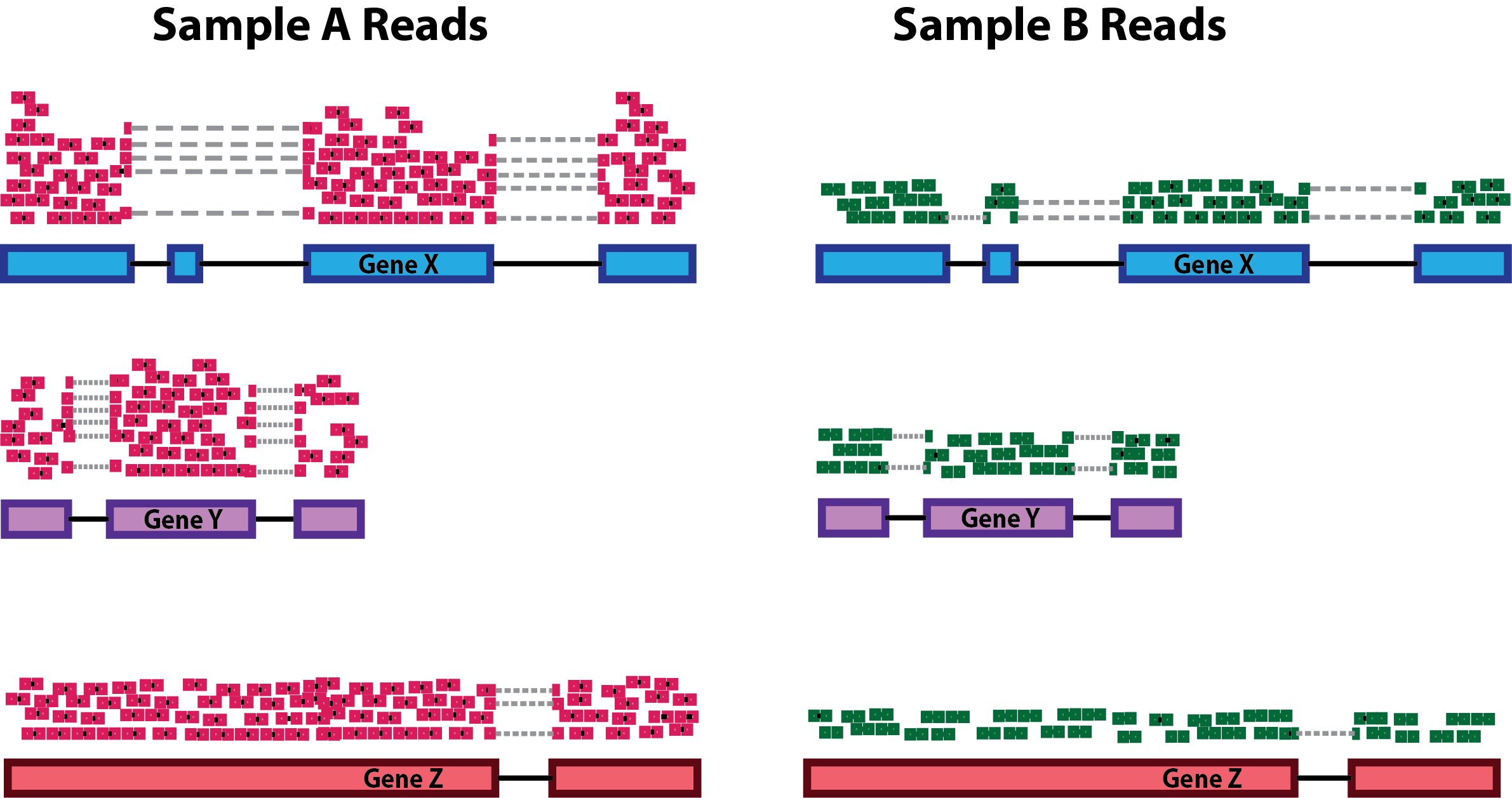

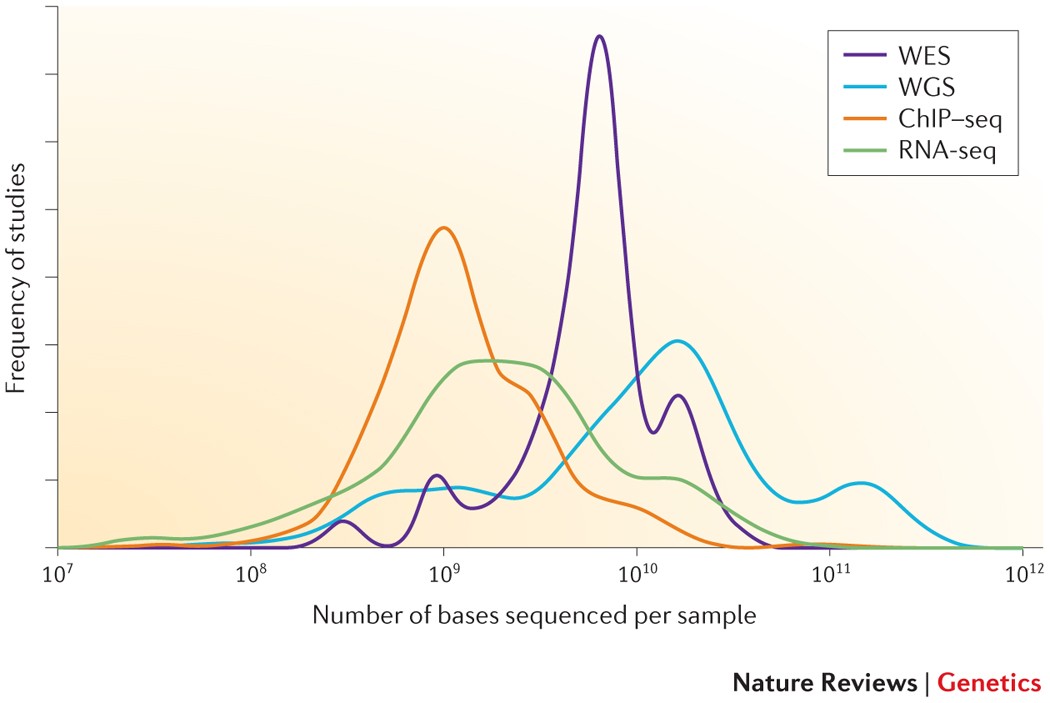
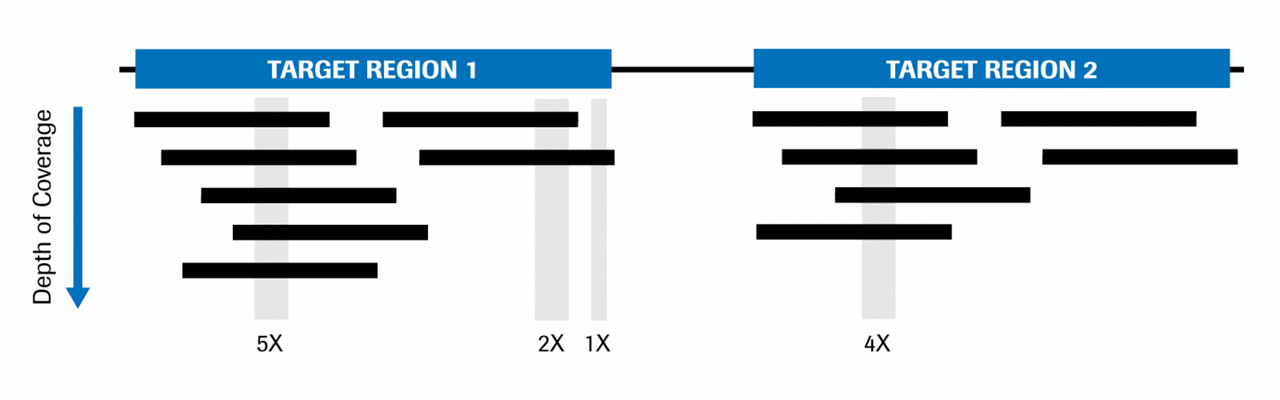








![PDF] Differential expression in RNA-seq: a matter of depth. | Semantic Scholar PDF] Differential expression in RNA-seq: a matter of depth. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/2cd8dc8fe9e89af280eef7e5b00cecd73e564da9/5-Figure2-1.png)

