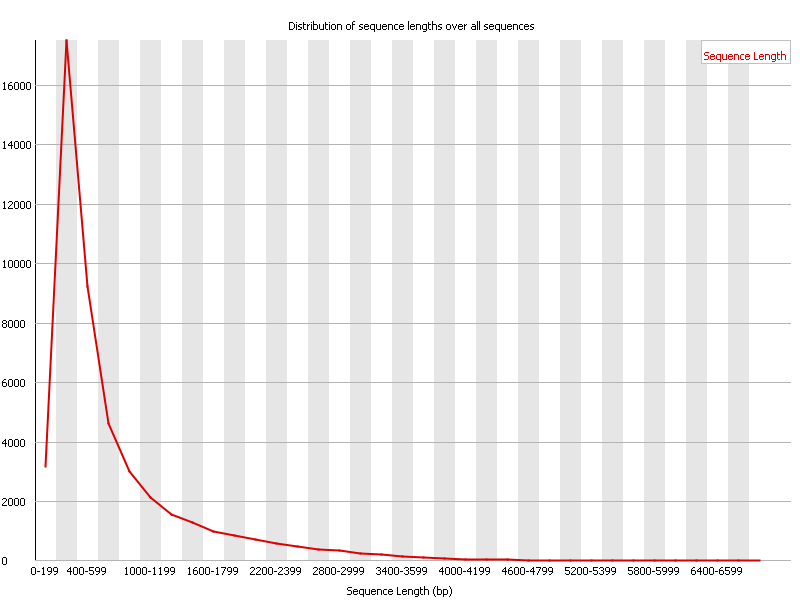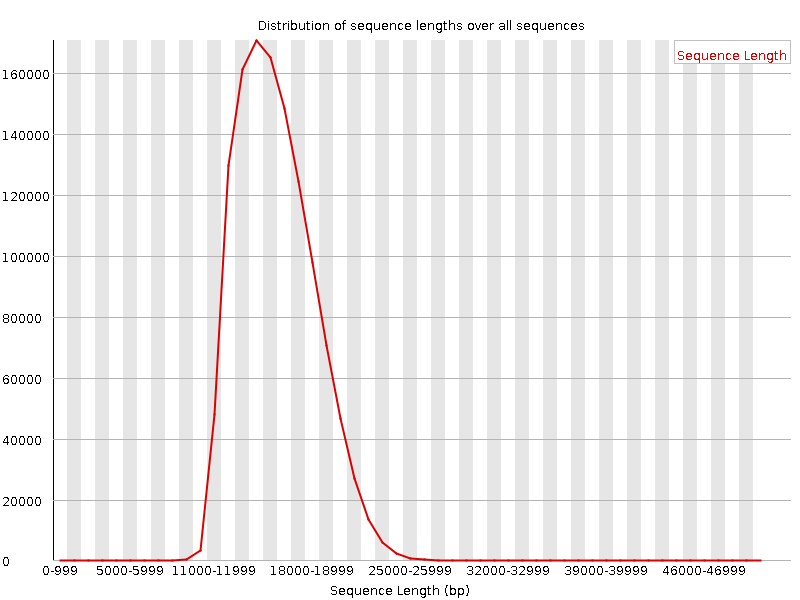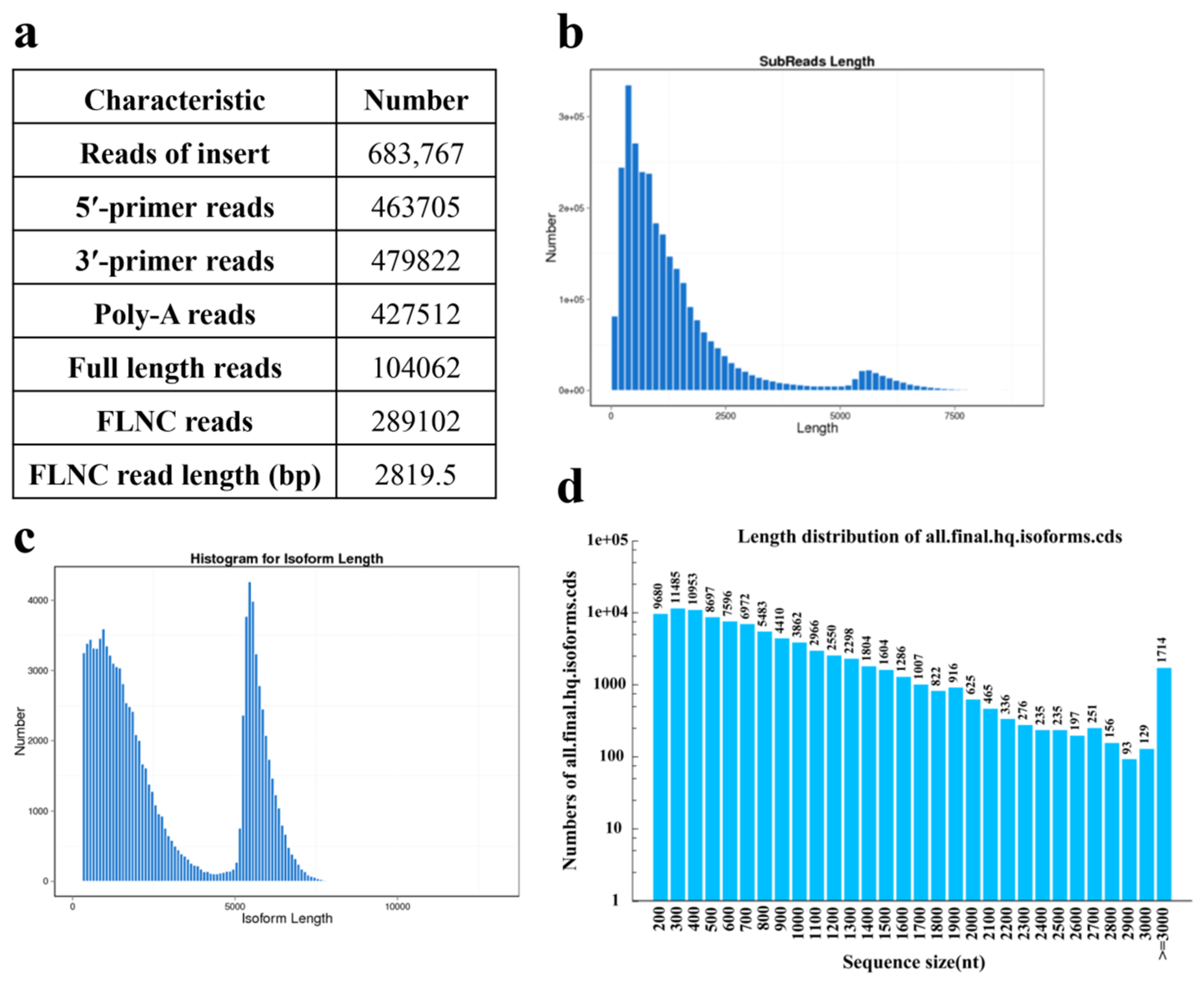
IJMS | Free Full-Text | Full-Length Transcriptome Sequencing and Different Chemotype Expression Profile Analysis of Genes Related to Monoterpenoid Biosynthesis in Cinnamomum porrectum
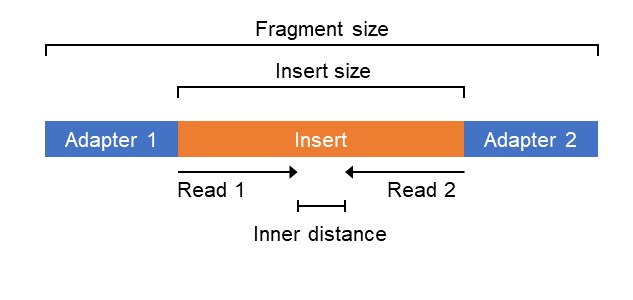
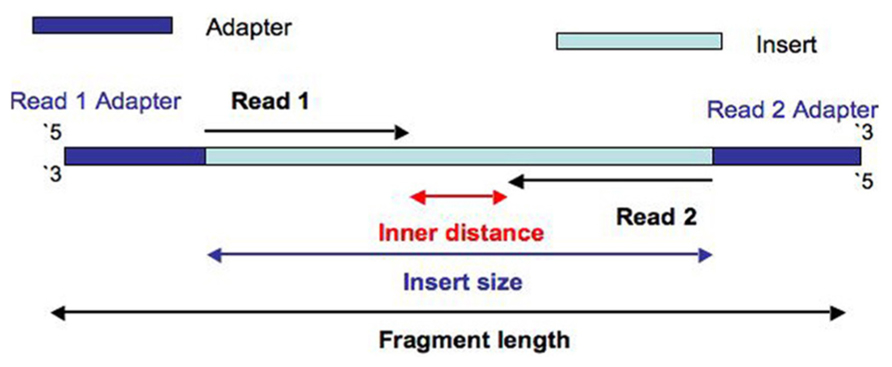
Compbio 020: Reads, fragments and inserts - what you need to know for understanding your sequencing data — Bad Grammar, Good Syntax

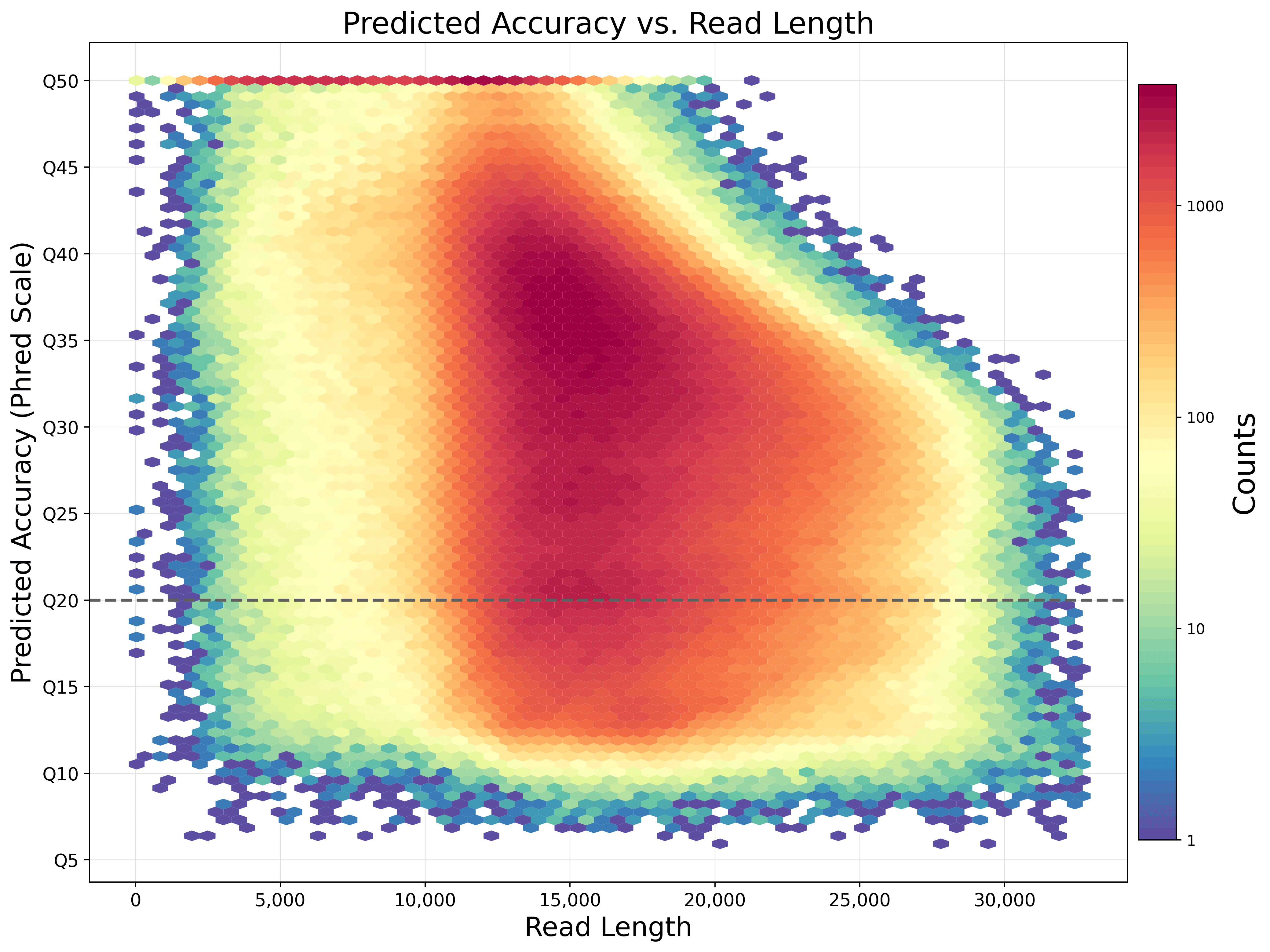
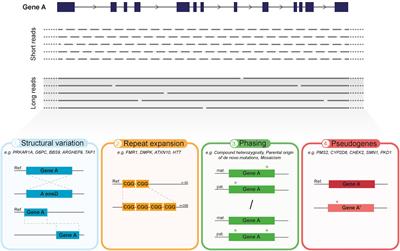
From kilobases to "whales": a short history of ultra-long reads and high-throughput genome sequencing