PepSeA: Peptide Sequence Alignment and Visualization Tools to Enable Lead Optimization | Journal of Chemical Information and Modeling
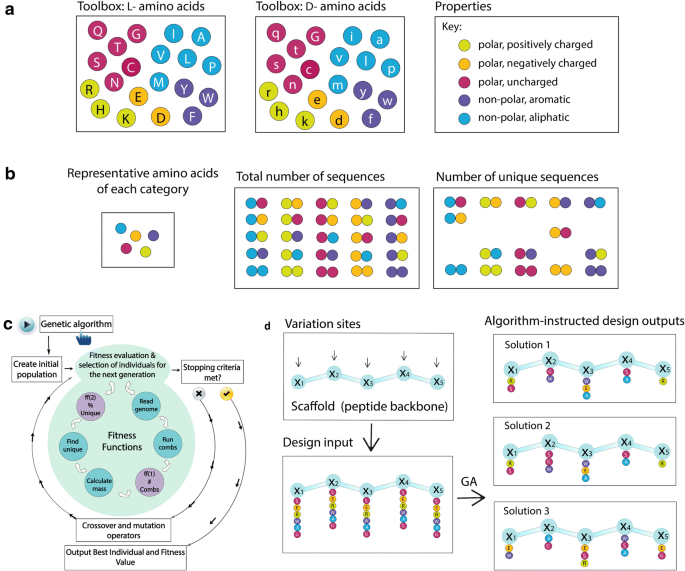
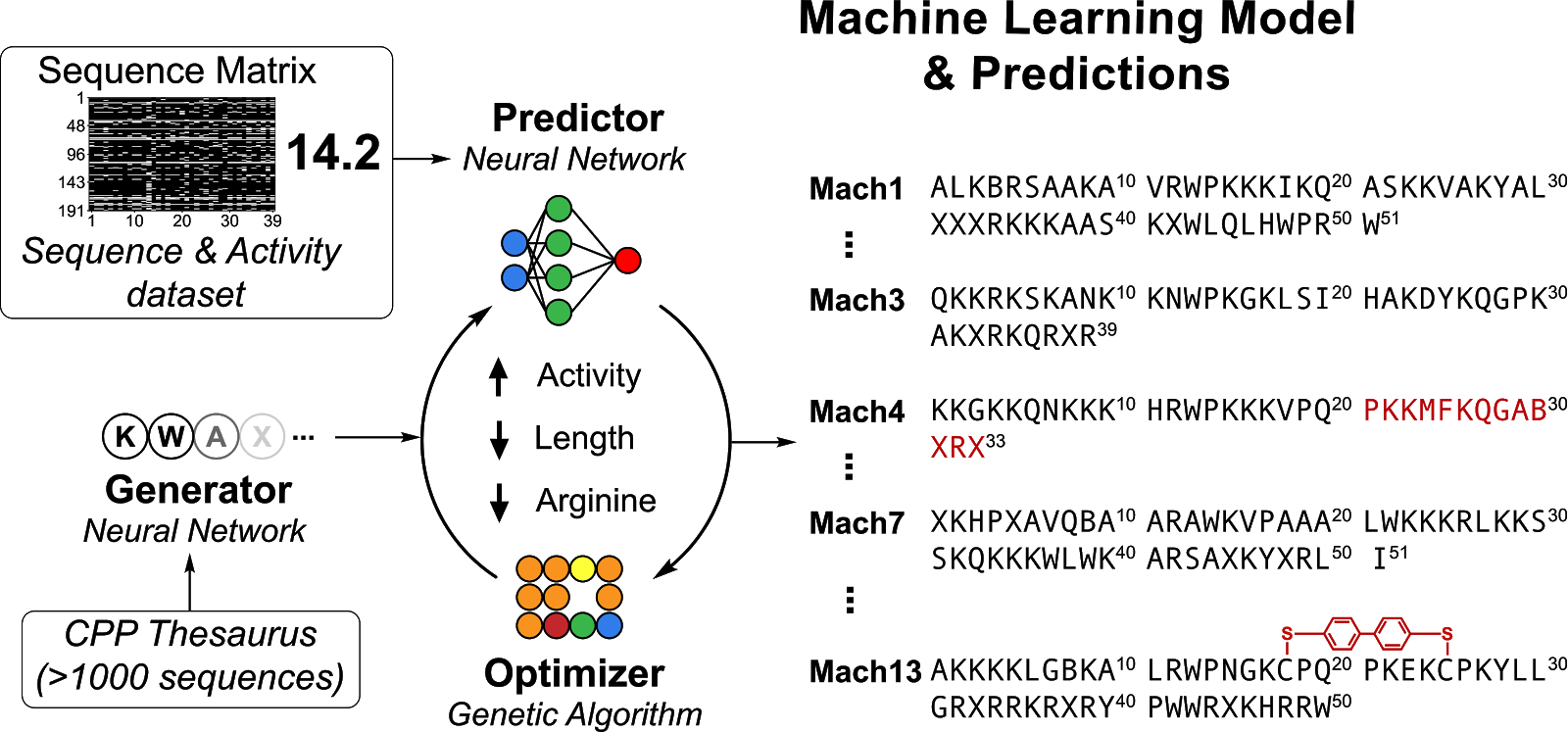
Algorithm-supported, mass and sequence diversity-oriented random peptide library design | Journal of Cheminformatics | Full Text

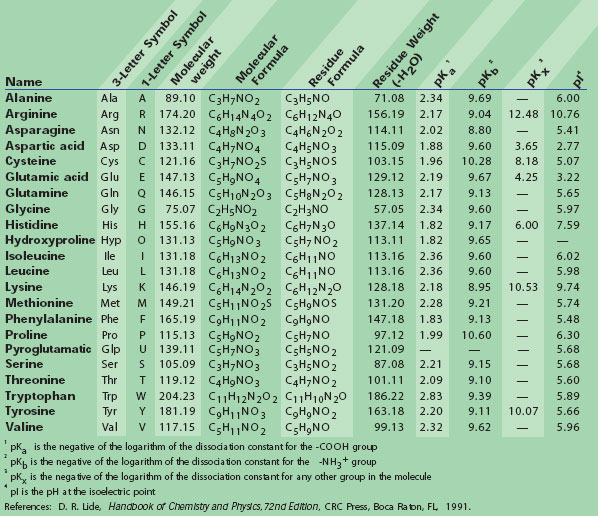
Stabilization Strategies for Linear Minigastrin Analogues: Further Improvements via the Inclusion of Proline into the Peptide Sequence | Journal of Medicinal Chemistry
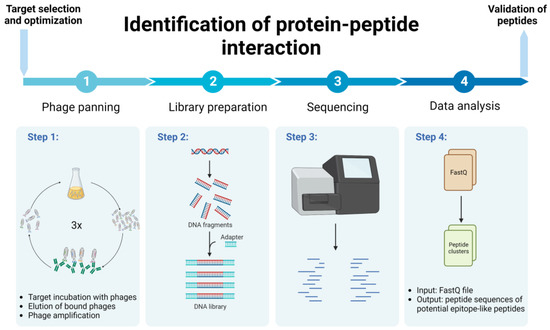
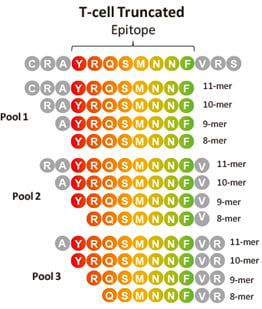
Biomolecules | Free Full-Text | Combination of Experimental and Bioinformatic Approaches for Identification of Immunologically Relevant Protein–Peptide Interactions

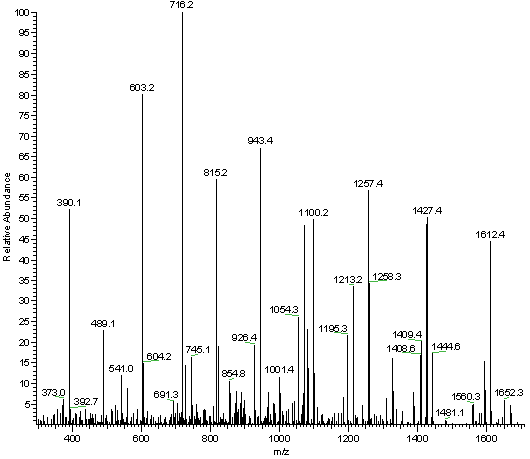
Uncovering Thousands of New Peptides with Sequence-Mask-Search Hybrid De Novo Peptide Sequencing Framework - ScienceDirect
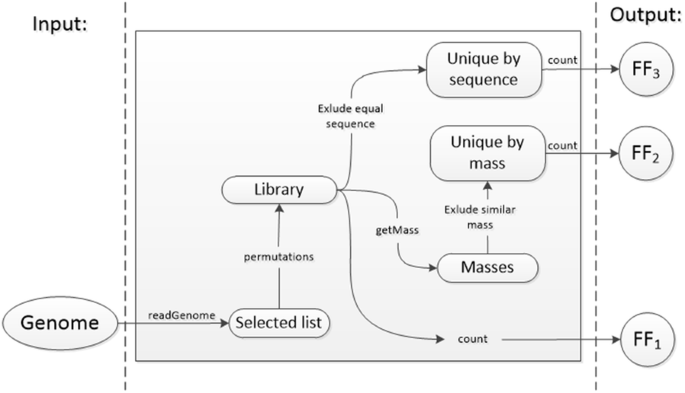
Algorithm-supported, mass and sequence diversity-oriented random peptide library design | Journal of Cheminformatics | Full Text

ADAPTABLE: a comprehensive web platform of antimicrobial peptides tailored to the user's research | Life Science Alliance
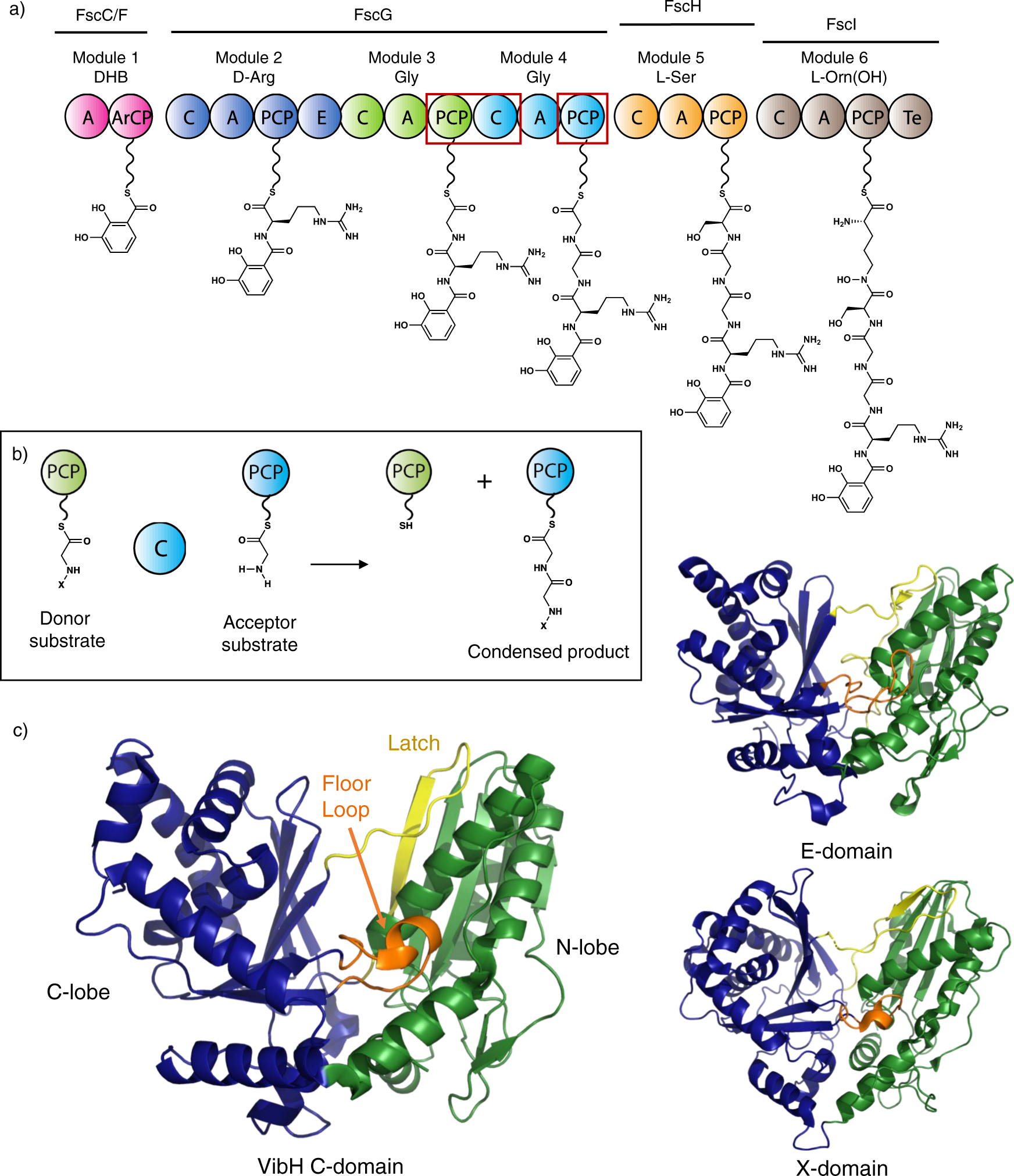
Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity | Nature Communications

PepSeA: Peptide Sequence Alignment and Visualization Tools to Enable Lead Optimization | Journal of Chemical Information and Modeling
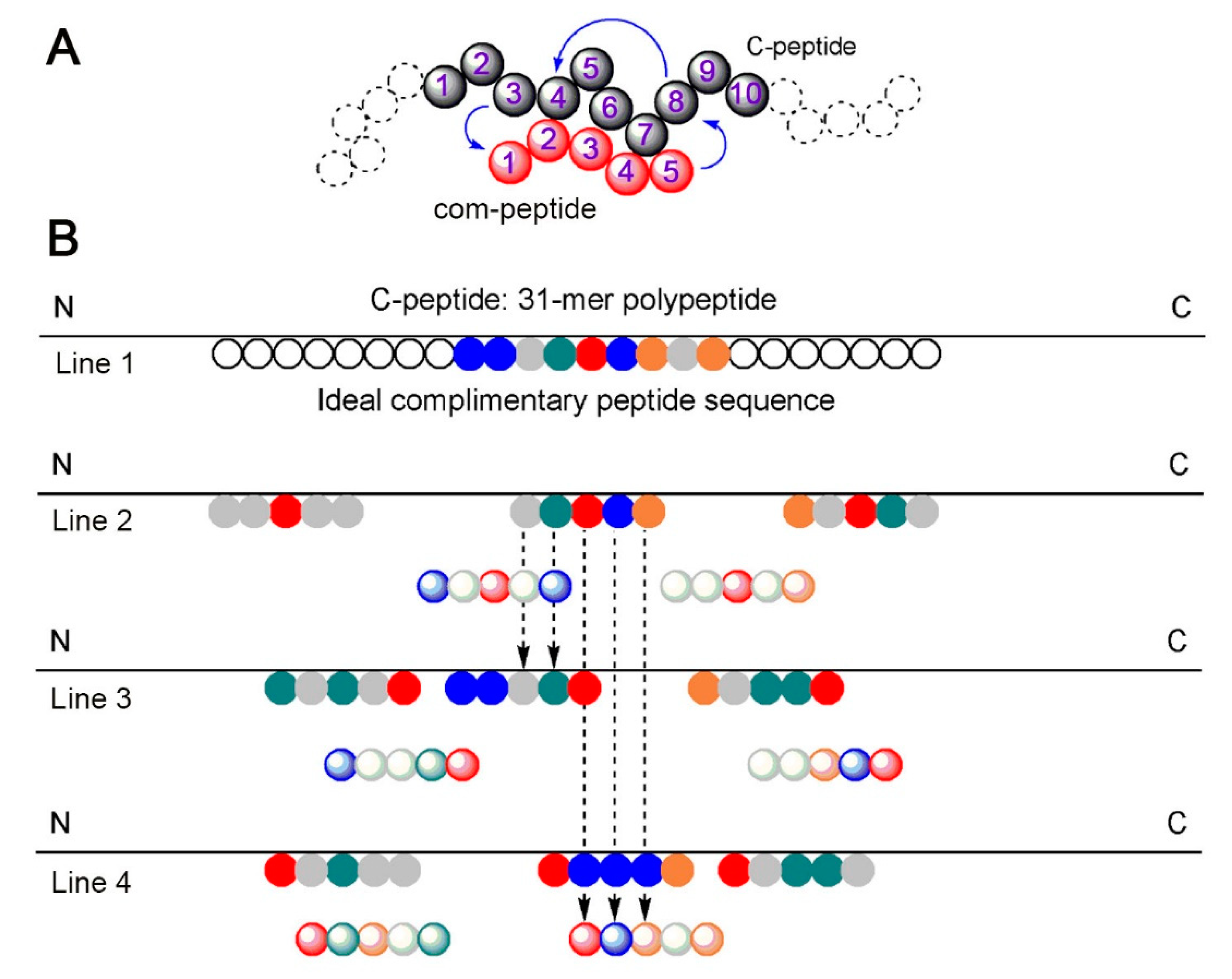
IJMS | Free Full-Text | Peptide–Peptide Co-Assembly: A Design Strategy for Functional Detection of C-peptide, A Biomarker of Diabetic Neuropathy
![PDF] PseKRAAC: a flexible web server for generating pseudo K-tuple reduced amino acids composition | Semantic Scholar PDF] PseKRAAC: a flexible web server for generating pseudo K-tuple reduced amino acids composition | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/bde9663b6cac60d4380e3827c0257604b29a8527/2-Table1-1.png)