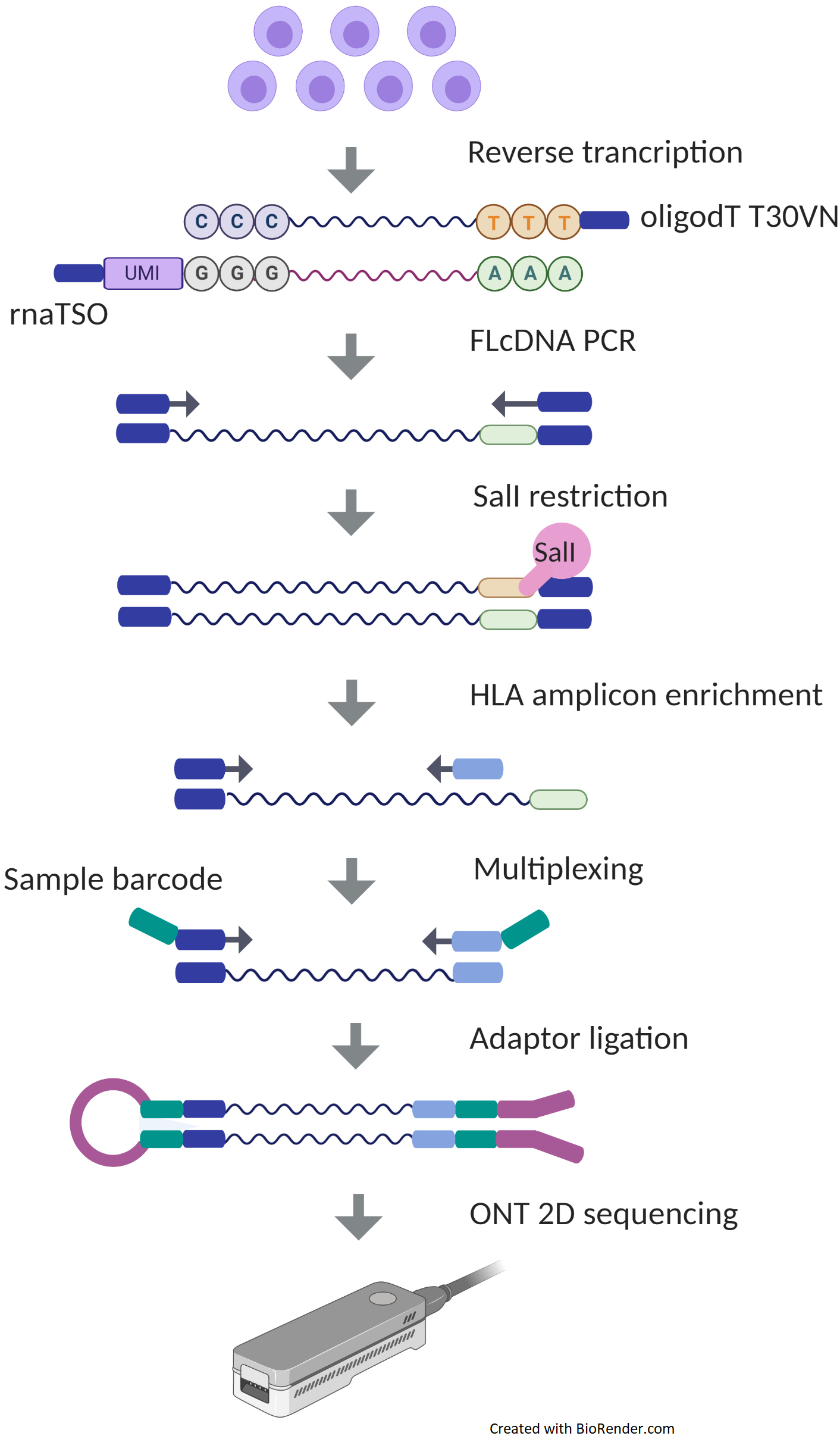
![Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis [PeerJ] Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/10778/1/fig-1-full.png)
Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis [PeerJ]

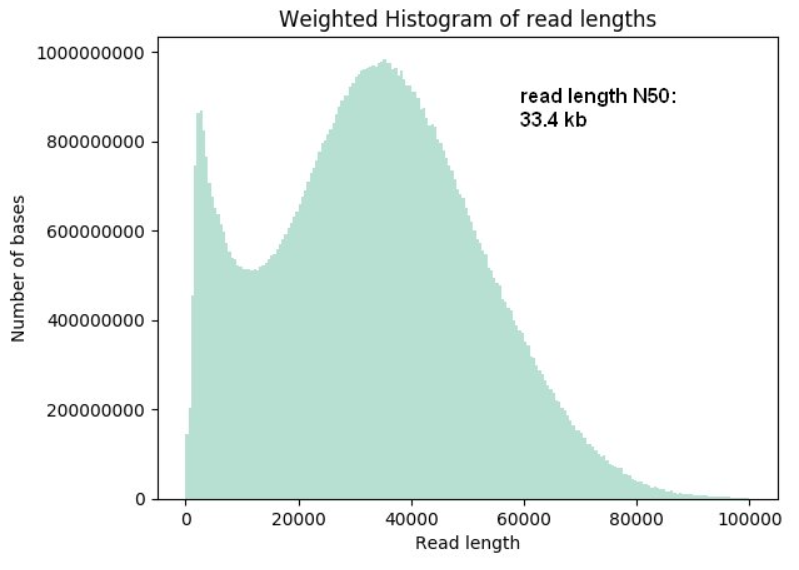
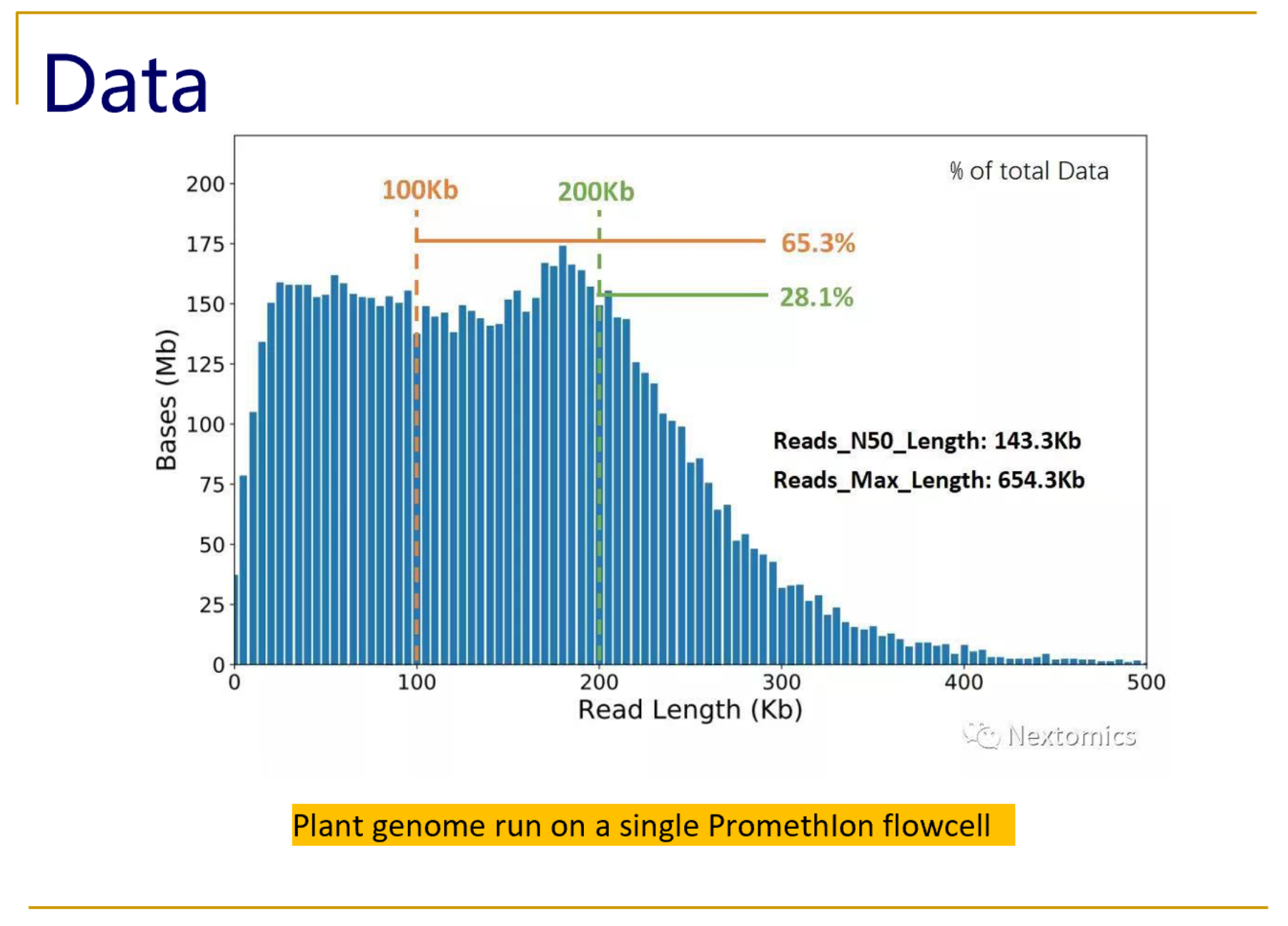
Nanopore read length distribution. Plot from the MinKNOW run report... | Download Scientific Diagram
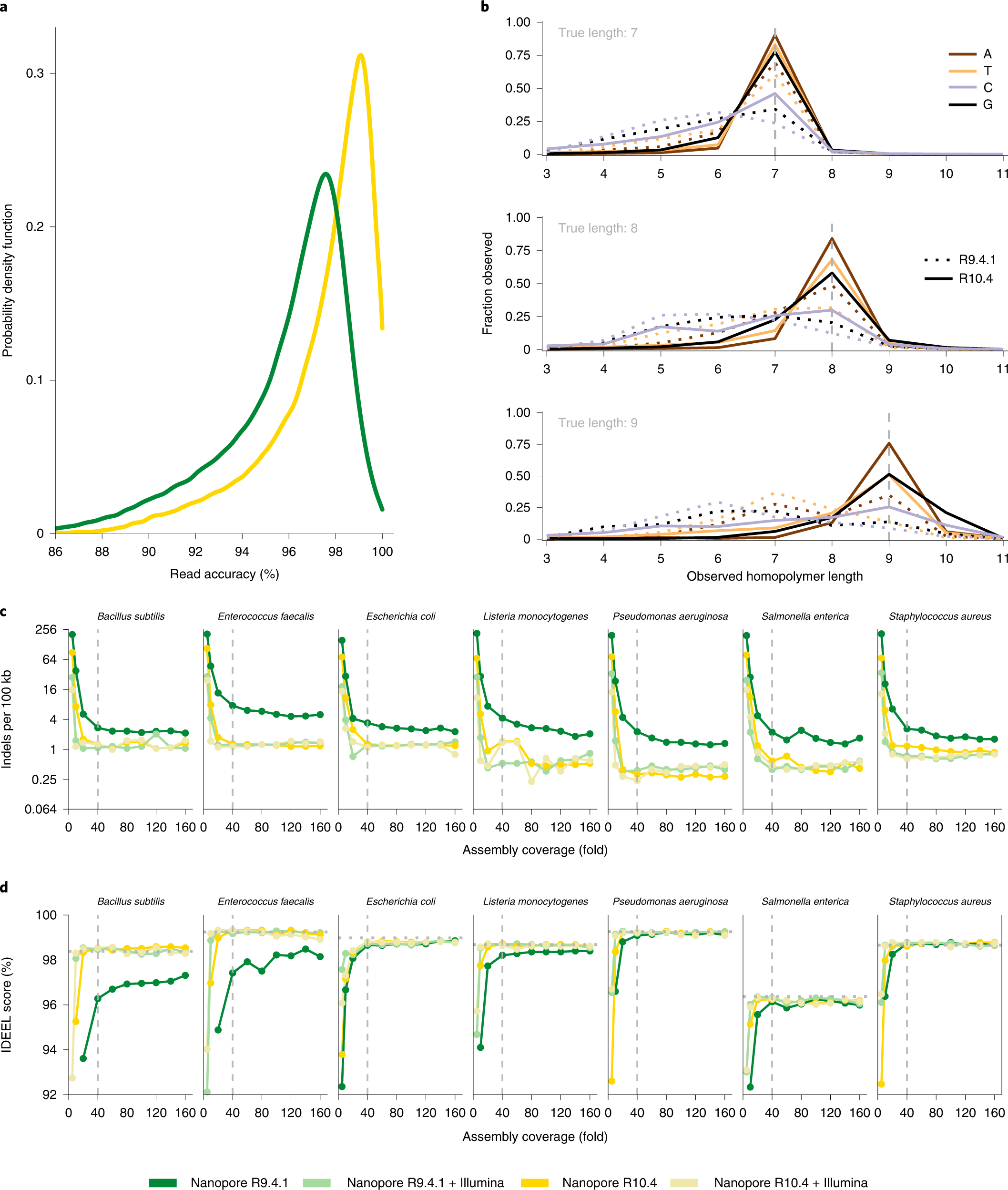
Oxford Nanopore R10.4 long-read sequencing enables the generation of near-finished bacterial genomes from pure cultures and metagenomes without short-read or reference polishing | Nature Methods
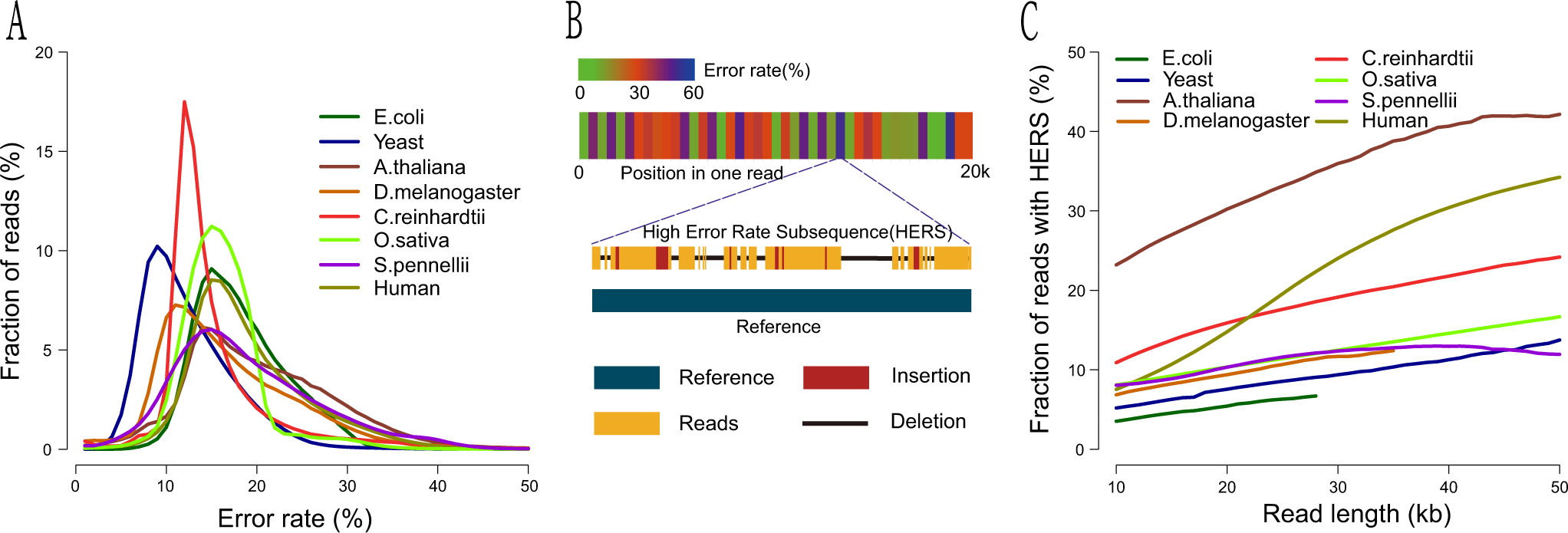
Efficient assembly of nanopore reads via highly accurate and intact error correction | Nature Communications