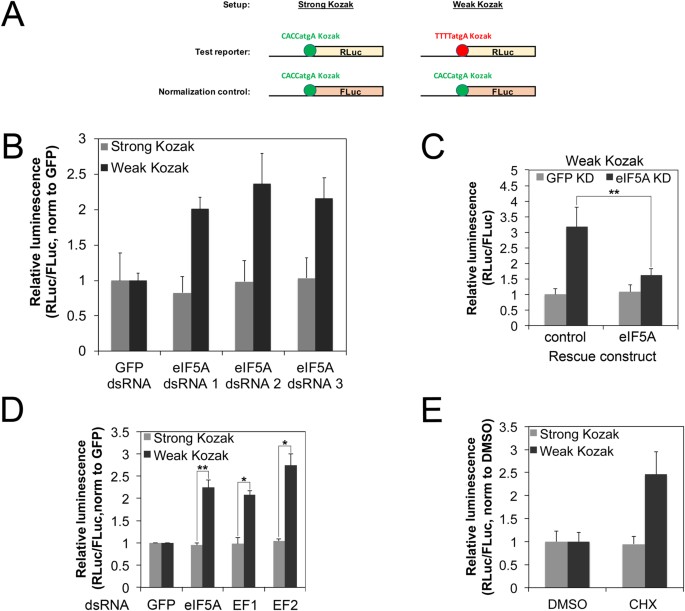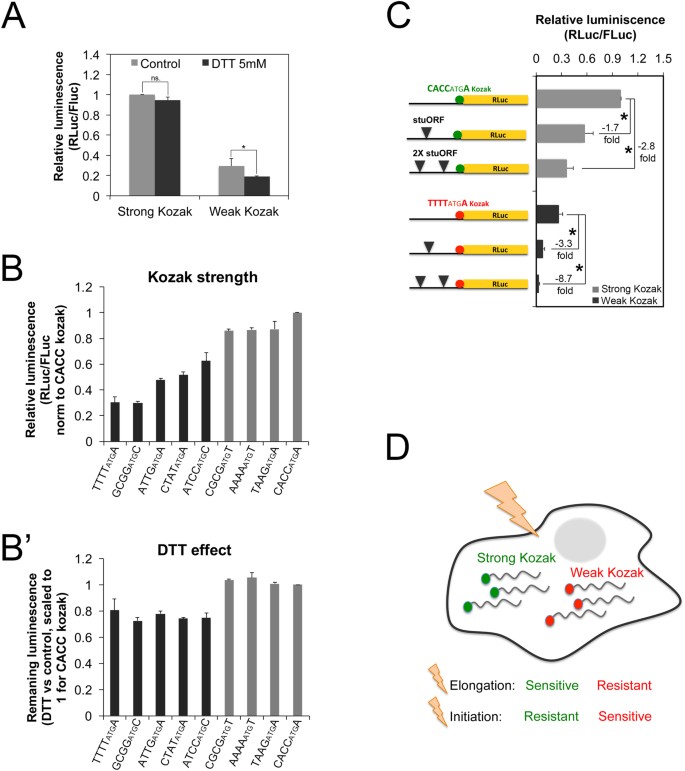
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports
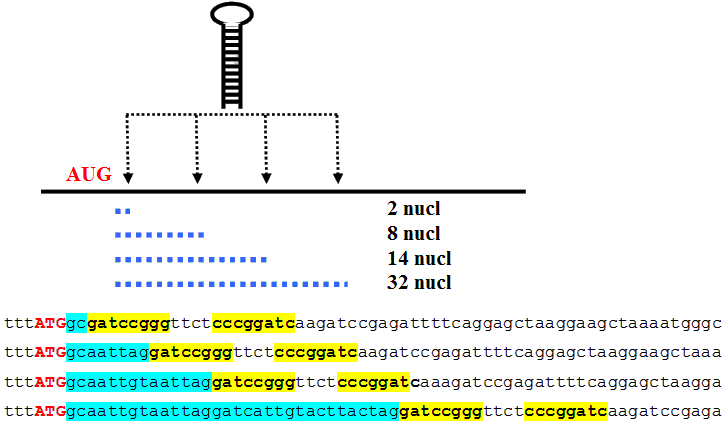
AUG_hairpin: program for prediction of a downstream hairpin potentially increasing initiation of translation at start AUG codon in a suboptimal context.
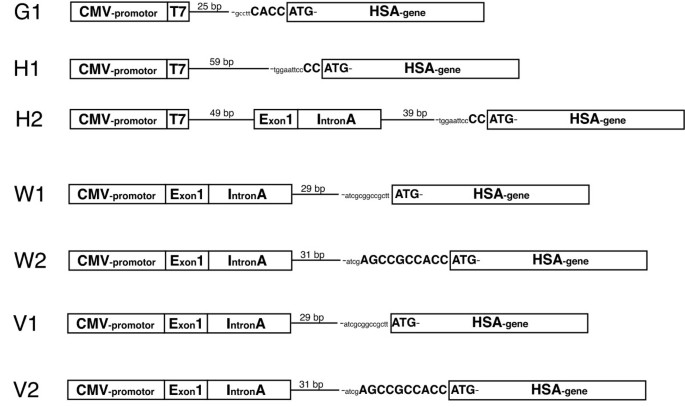
In vitro analysis of expression vectors for DNA vaccination of horses: the effect of a Kozak sequence | Acta Veterinaria Scandinavica | Full Text

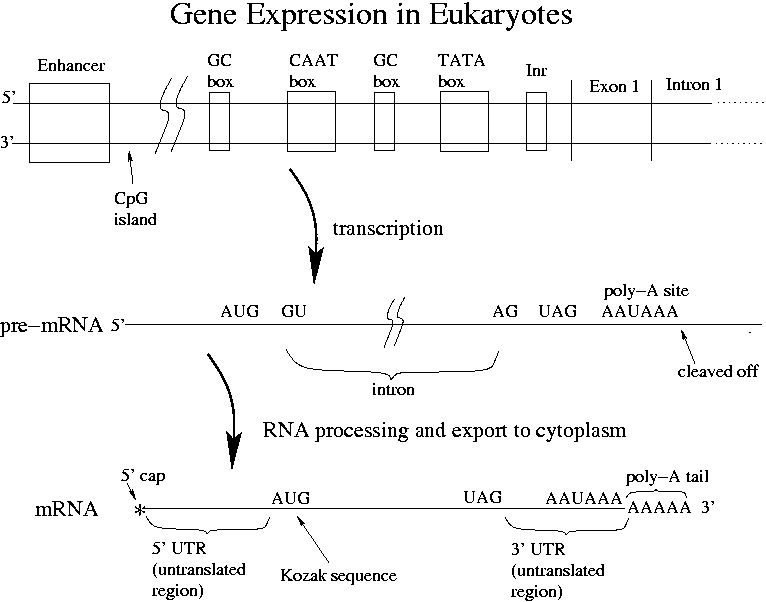
Quantitative analysis of mammalian translation initiation sites by FACS‐seq | Molecular Systems Biology

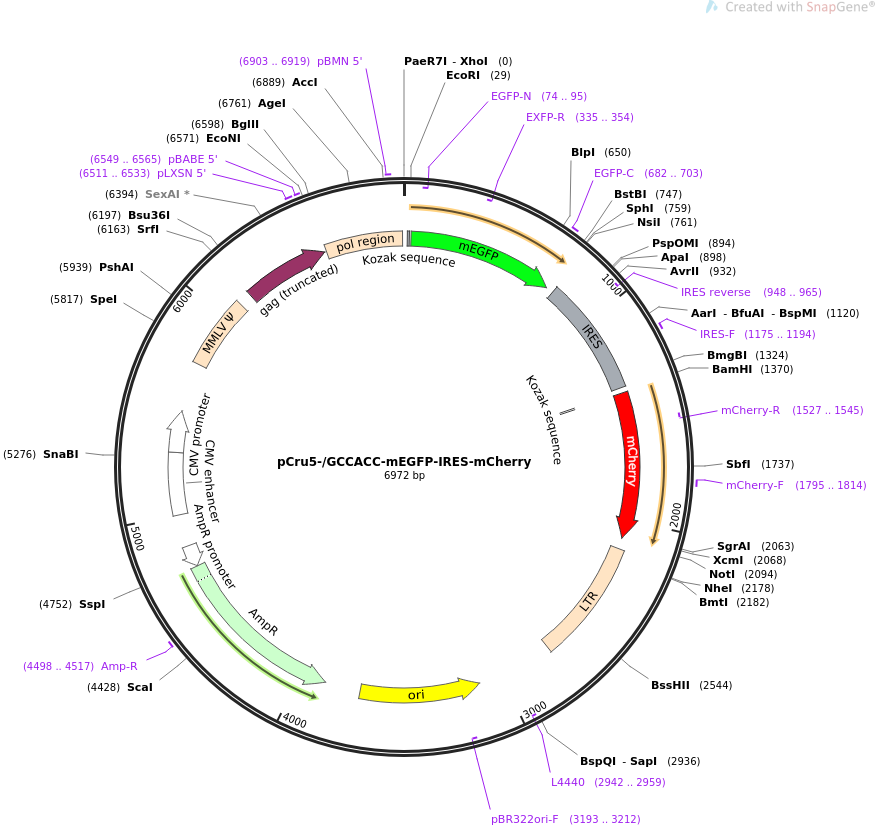
Combination of Kozak sequence and optimized gene arrangement further... | Download Scientific Diagram
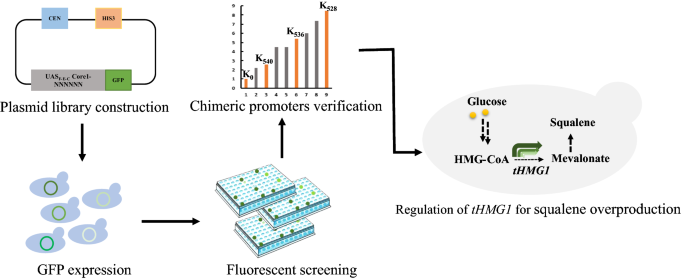
Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants | Microbial Cell Factories | Full Text

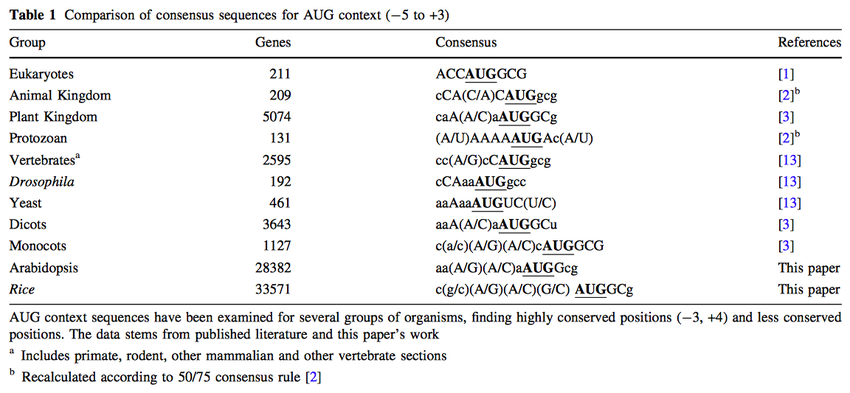
Conservation and Variability of the AUG Initiation Codon Context in Eukaryotes: Trends in Biochemical Sciences

Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants | Microbial Cell Factories | Full Text

Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv
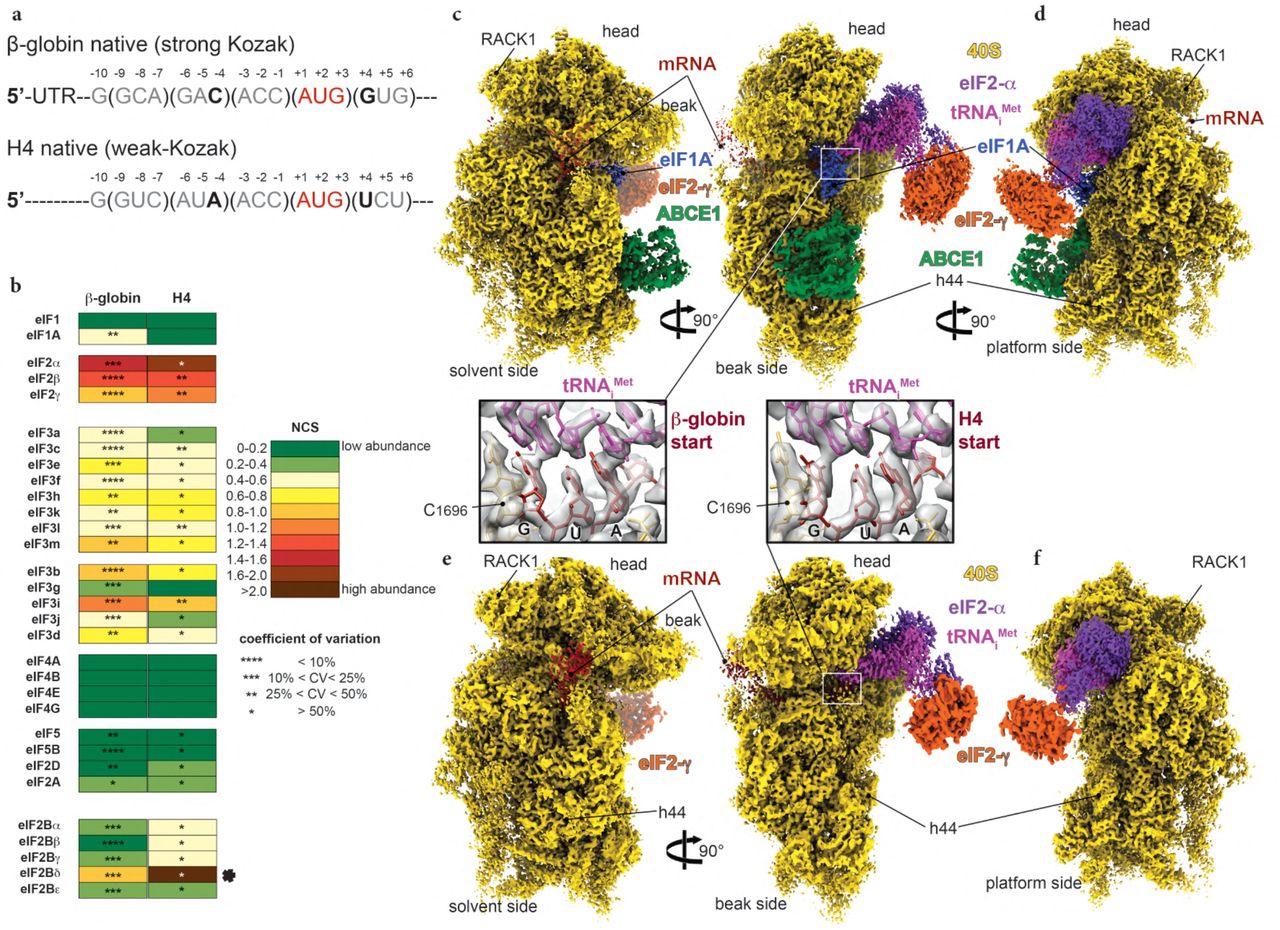
Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv