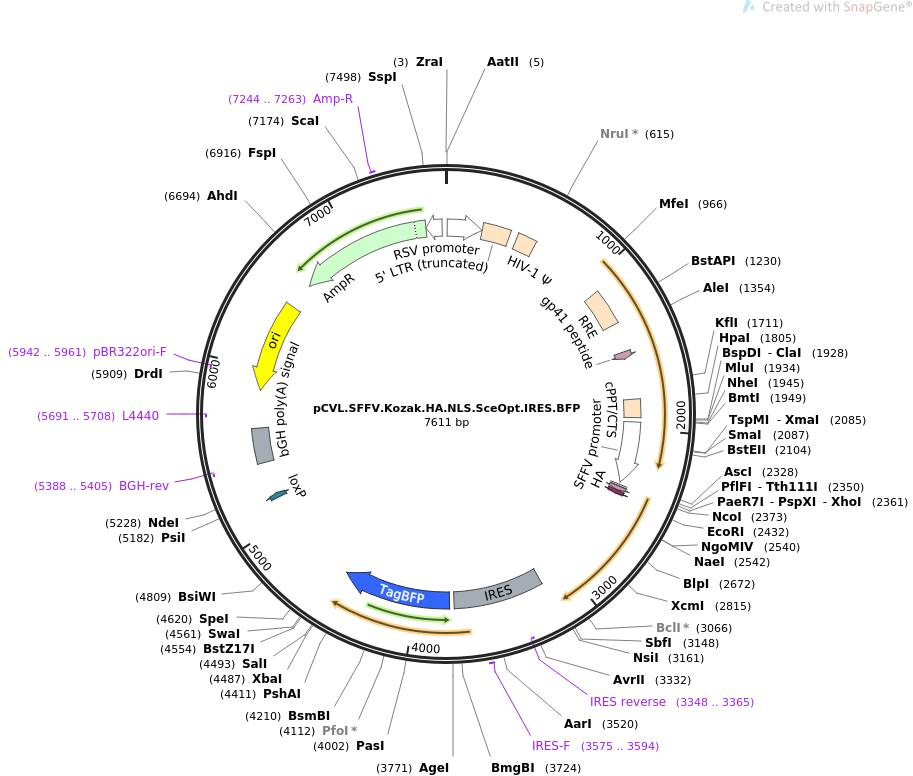
Heterologous expression and cell membrane localization of dinoflagellate opsins (rhodopsin proteins) in mammalian cells | SpringerLink

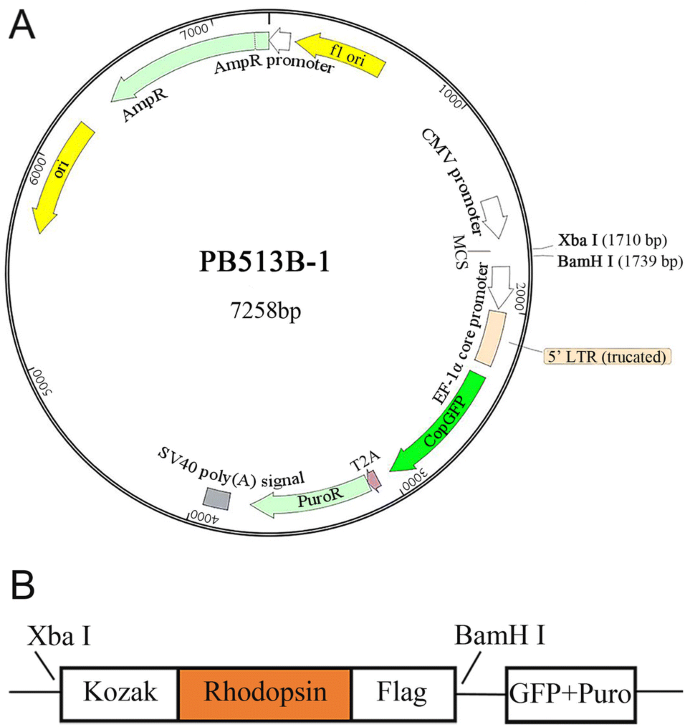
Combination of Kozak sequence and optimized gene arrangement further... | Download Scientific Diagram

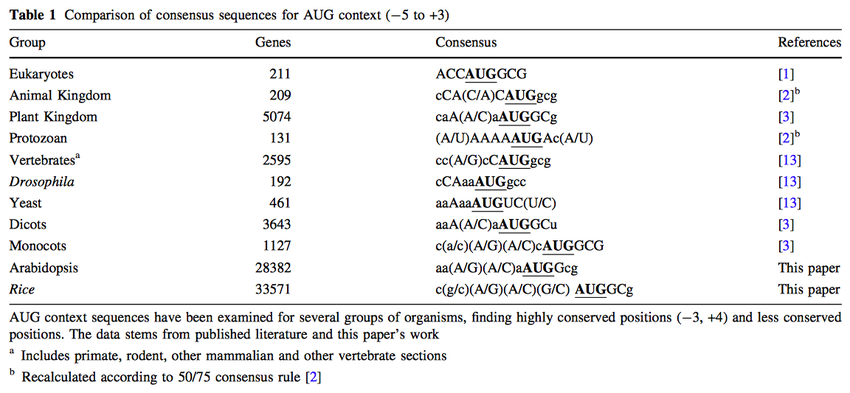
Conservation and Variability of the AUG Initiation Codon Context in Eukaryotes: Trends in Biochemical Sciences
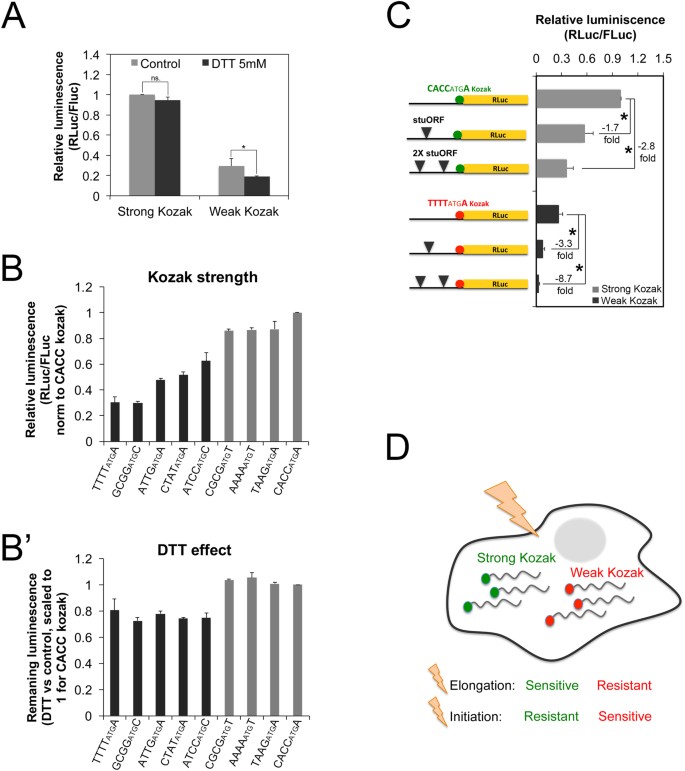
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports
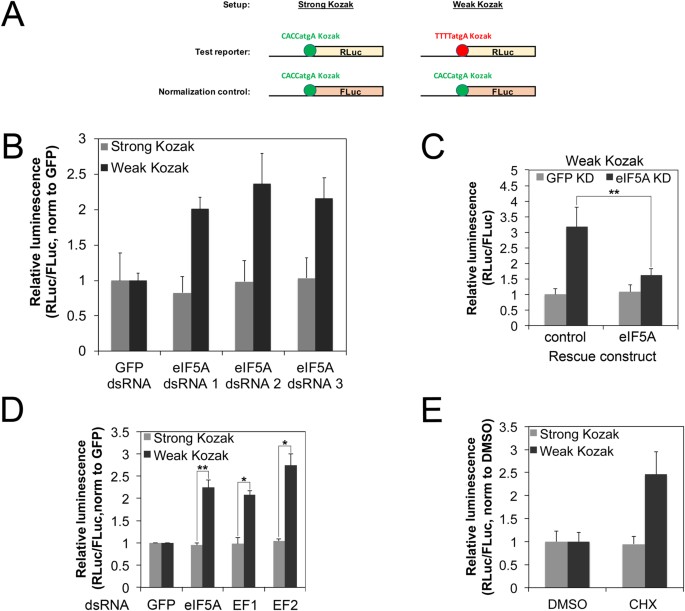
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports

Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv

Kozak sequence improved the expression of 2A downstream gene by its... | Download Scientific Diagram
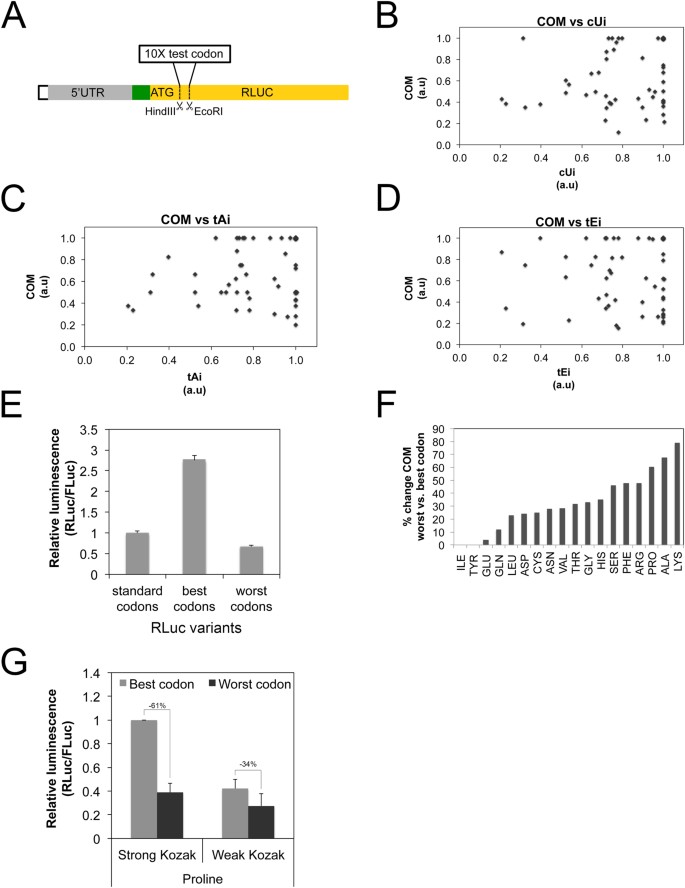
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports

IRES-Dependent Second Gene Expression Is Significantly Lower Than Cap-Dependent First Gene Expression in a Bicistronic Vector: Molecular Therapy

Bioinformatic analyses of mammalian 5'-UTR sequence properties of mRNAs predicts alternative translation initiation sites | BMC Bioinformatics | Full Text

Nucleotides upstream of the Kozak sequence strongly influence gene expression in the yeast S. cerevisiae | Journal of Biological Engineering | Full Text
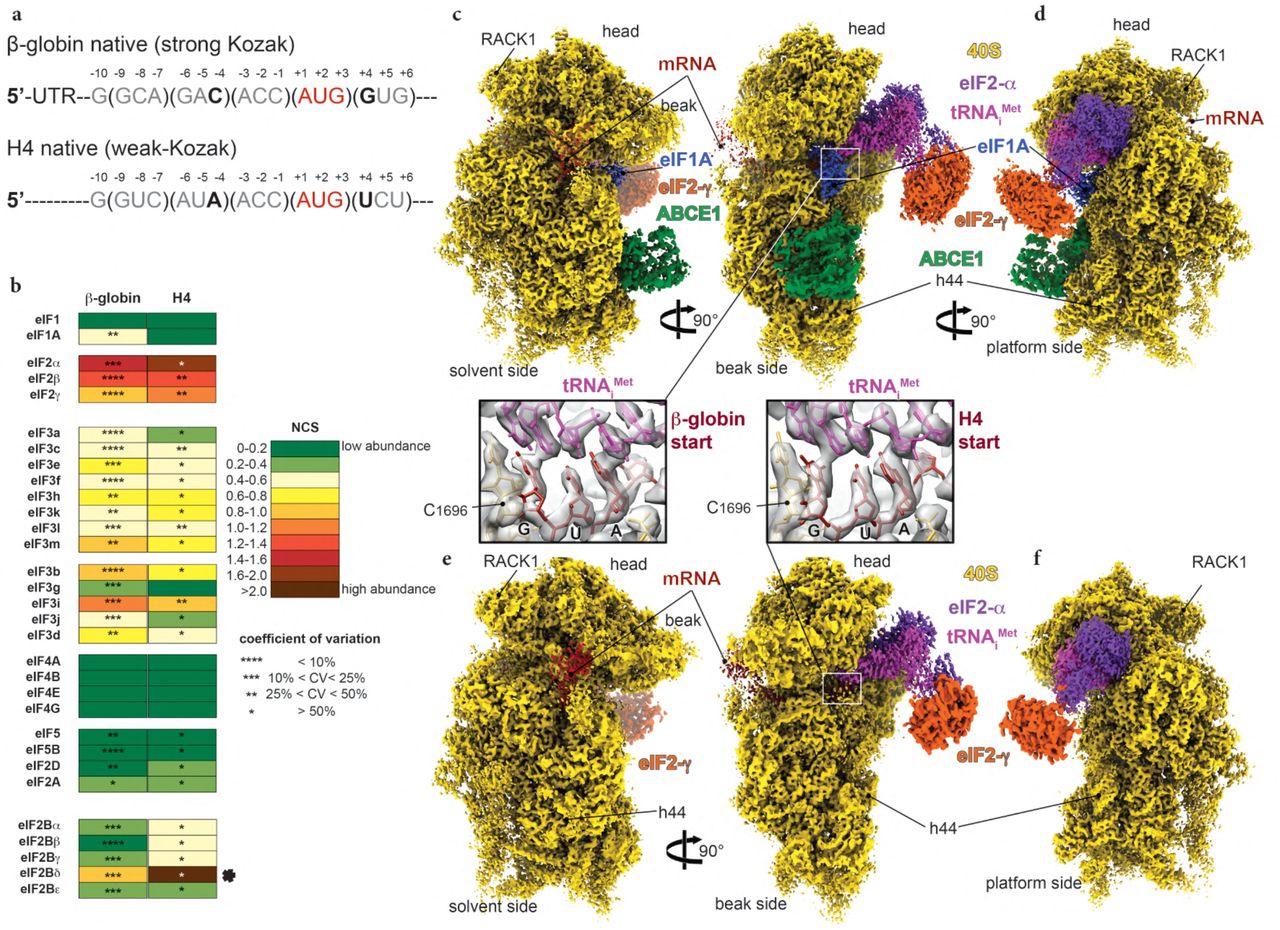
Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv

Enhancement of protein expression in insect cells by a lobster tropomyosin cDNA leader sequence - ScienceDirect