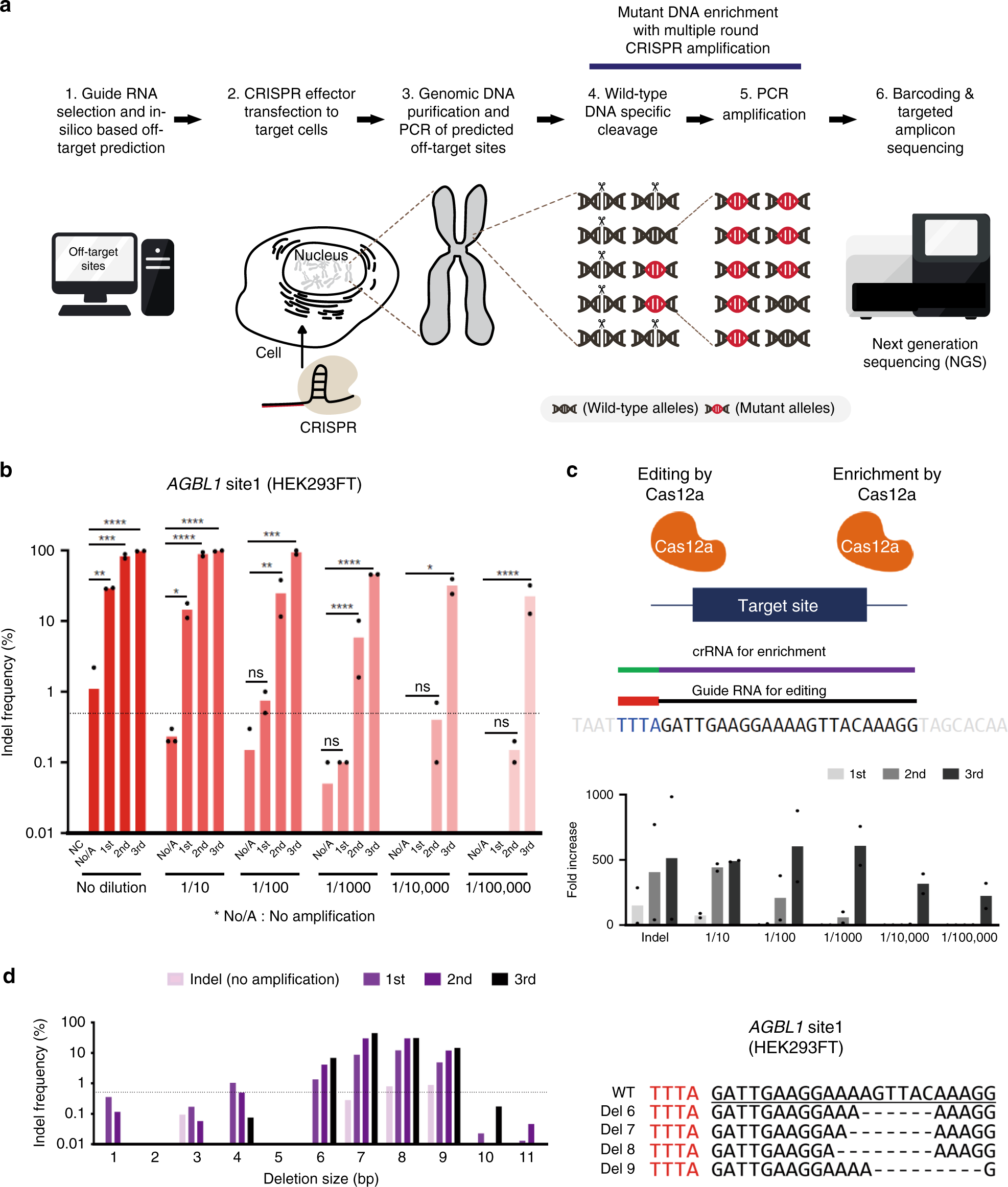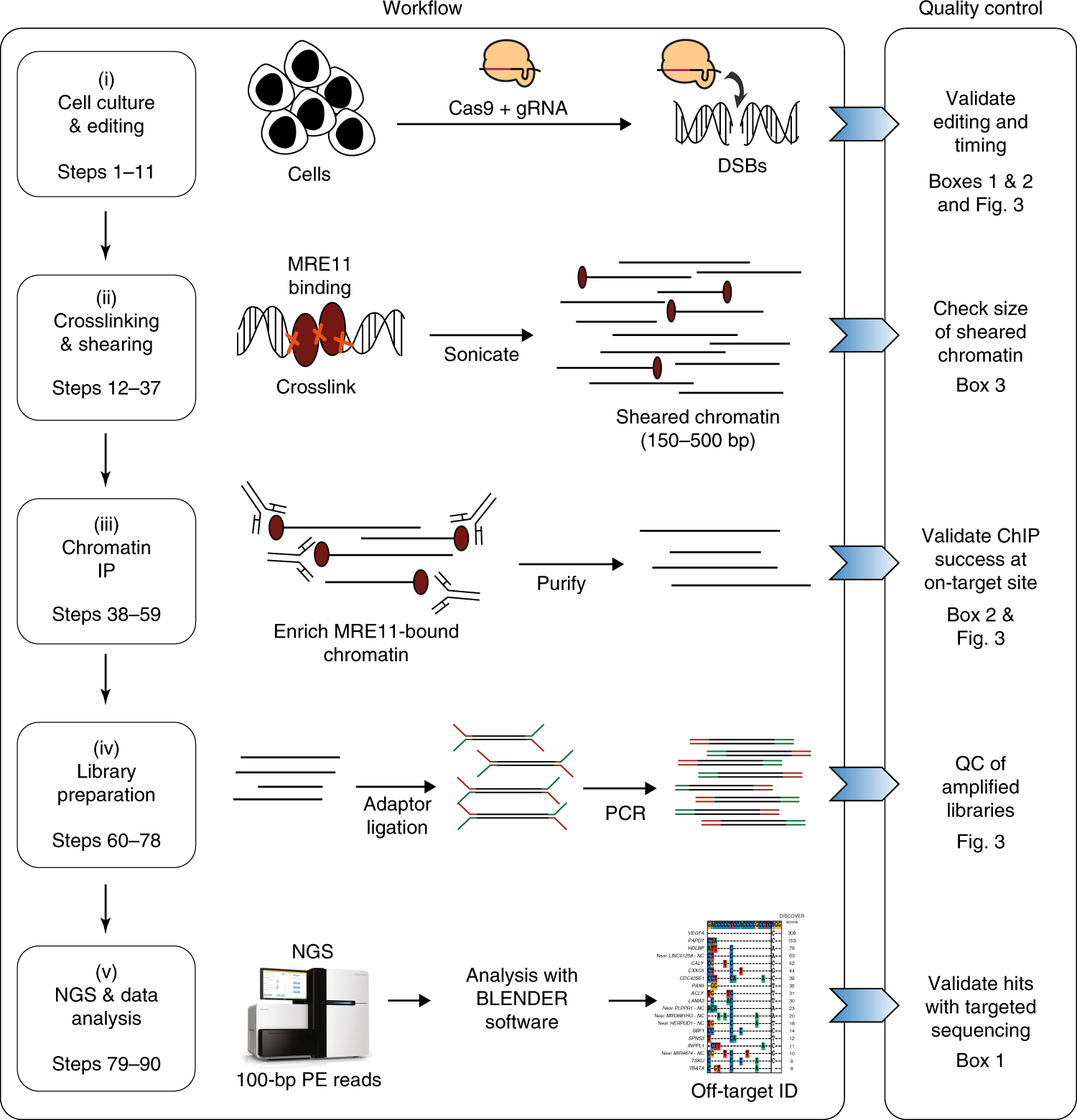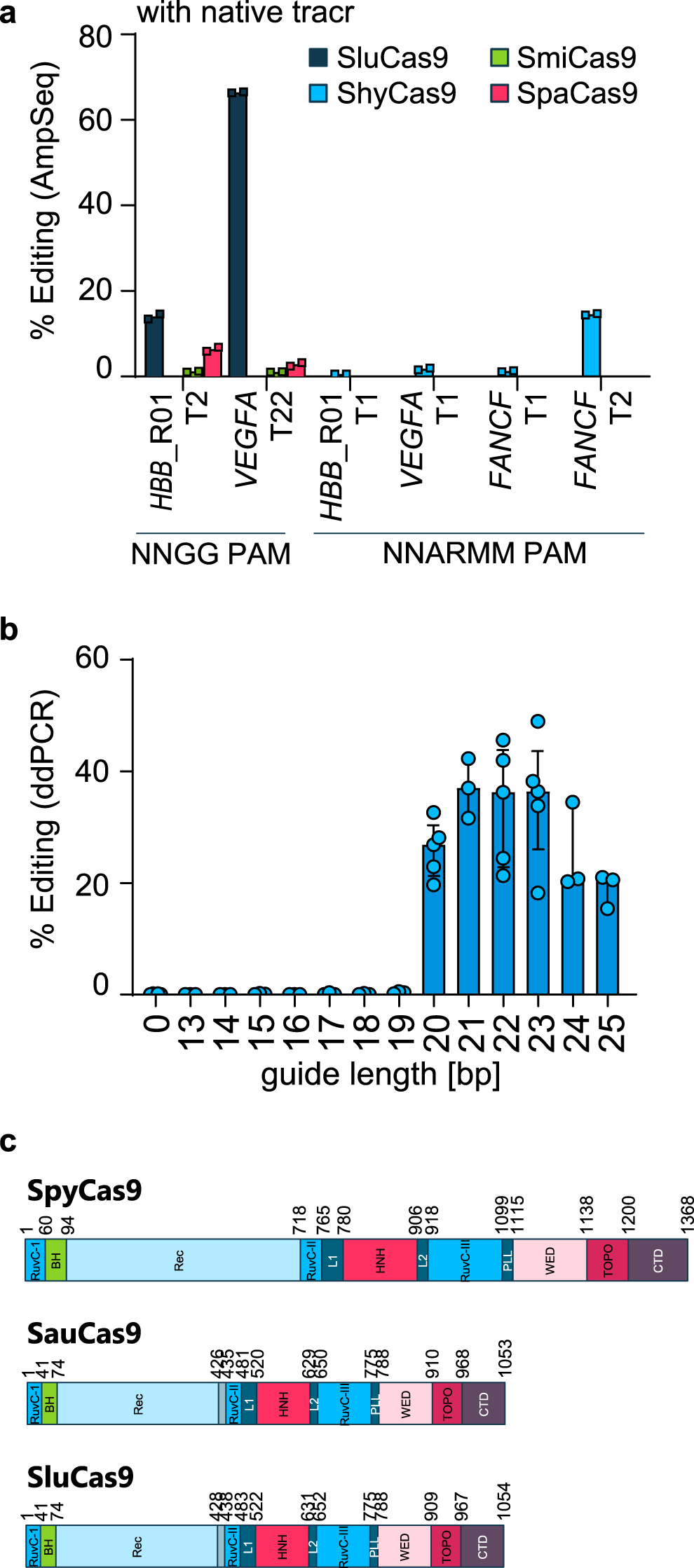
Improved CRISPR genome editing using small highly active and specific engineered RNA-guided nucleases | Nature Communications

Targeted Amplicon Sequencing for Single-Nucleotide-Polymorphism Genotyping of Attaching and Effacing Escherichia coli O26:H11 Cattle Strains via a High-Throughput Library Preparation Technique | Applied and Environmental Microbiology
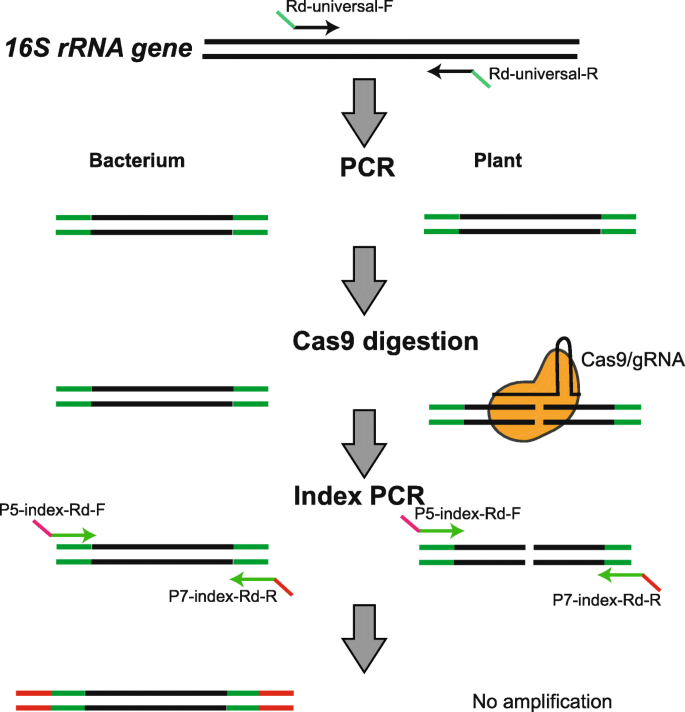
Engineering CRISPR/Cas9 to mitigate abundant host contamination for 16S rRNA gene-based amplicon sequencing | Microbiome | Full Text
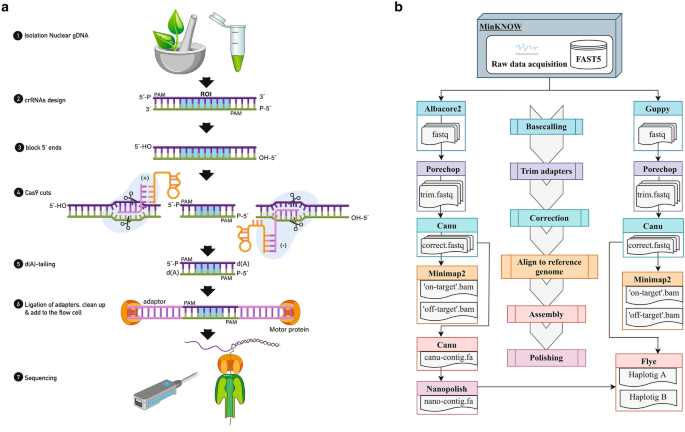
CRISPR-Cas9 enrichment and long read sequencing for fine mapping in plants | Plant Methods | Full Text
Techniques that use CRISPR-Cas systems to generate amplified signals... | Download Scientific Diagram

CRISPR-Detector: Fast and Accurate Detection, Visualization, and Annotation of Genome Wide Mutations Induced by Gene Editing Events | bioRxiv

A simple and practical workflow for genotyping of CRISPR–Cas9‐based knockout phenotypes using multiplexed amplicon sequencing - Iida - 2020 - Genes to Cells - Wiley Online Library
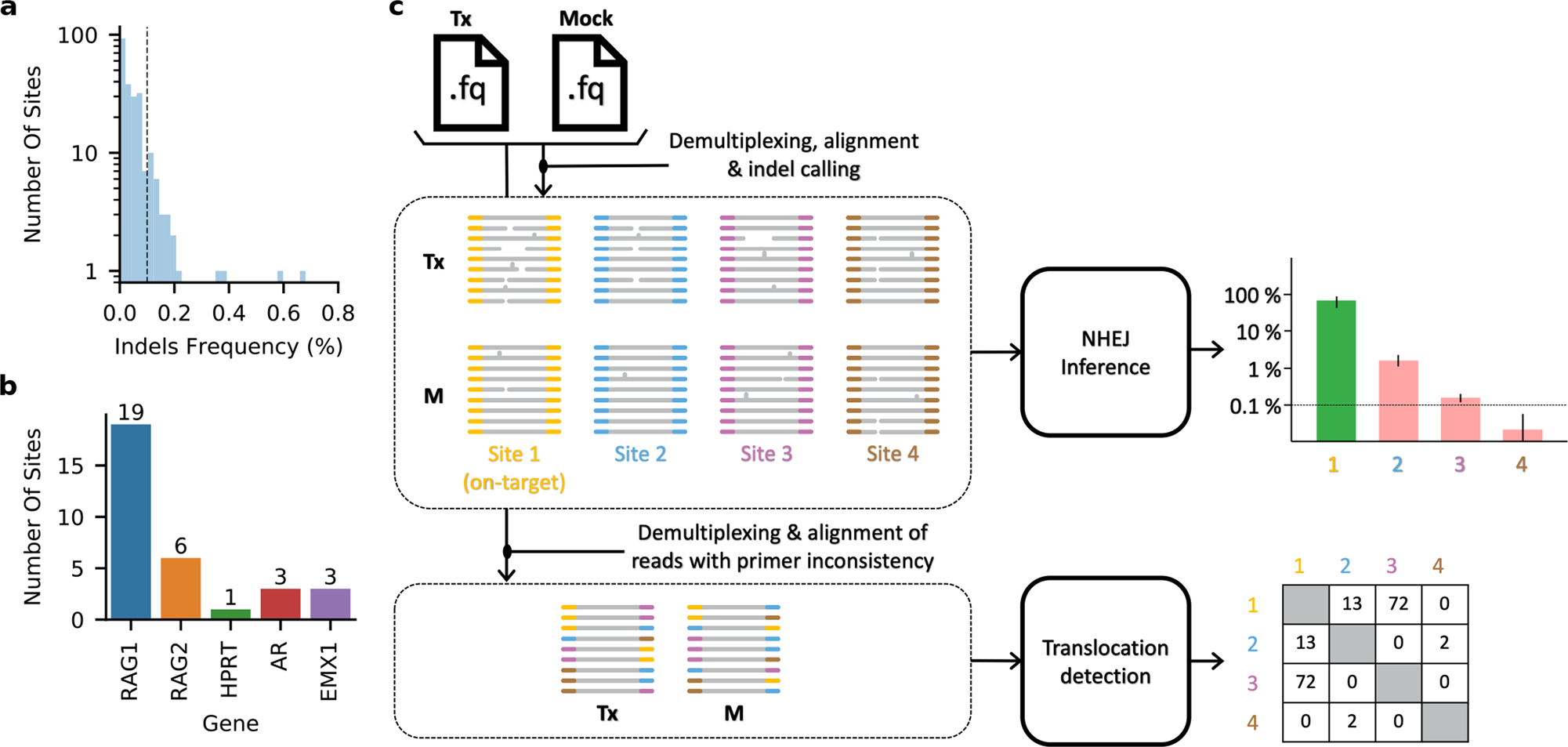
CRISPECTOR provides accurate estimation of genome editing translocation and off-target activity from comparative NGS data | Nature Communications

evSeq: Cost-Effective Amplicon Sequencing of Every Variant in a Protein Library | ACS Synthetic Biology

A simple and practical workflow for genotyping of CRISPR–Cas9‐based knockout phenotypes using multiplexed amplicon sequencing - Iida - 2020 - Genes to Cells - Wiley Online Library

The use of CRISPR-Cas Selective Amplicon Sequencing (CCSAS) to reveal the eukaryotic microbiome of metazoans | bioRxiv
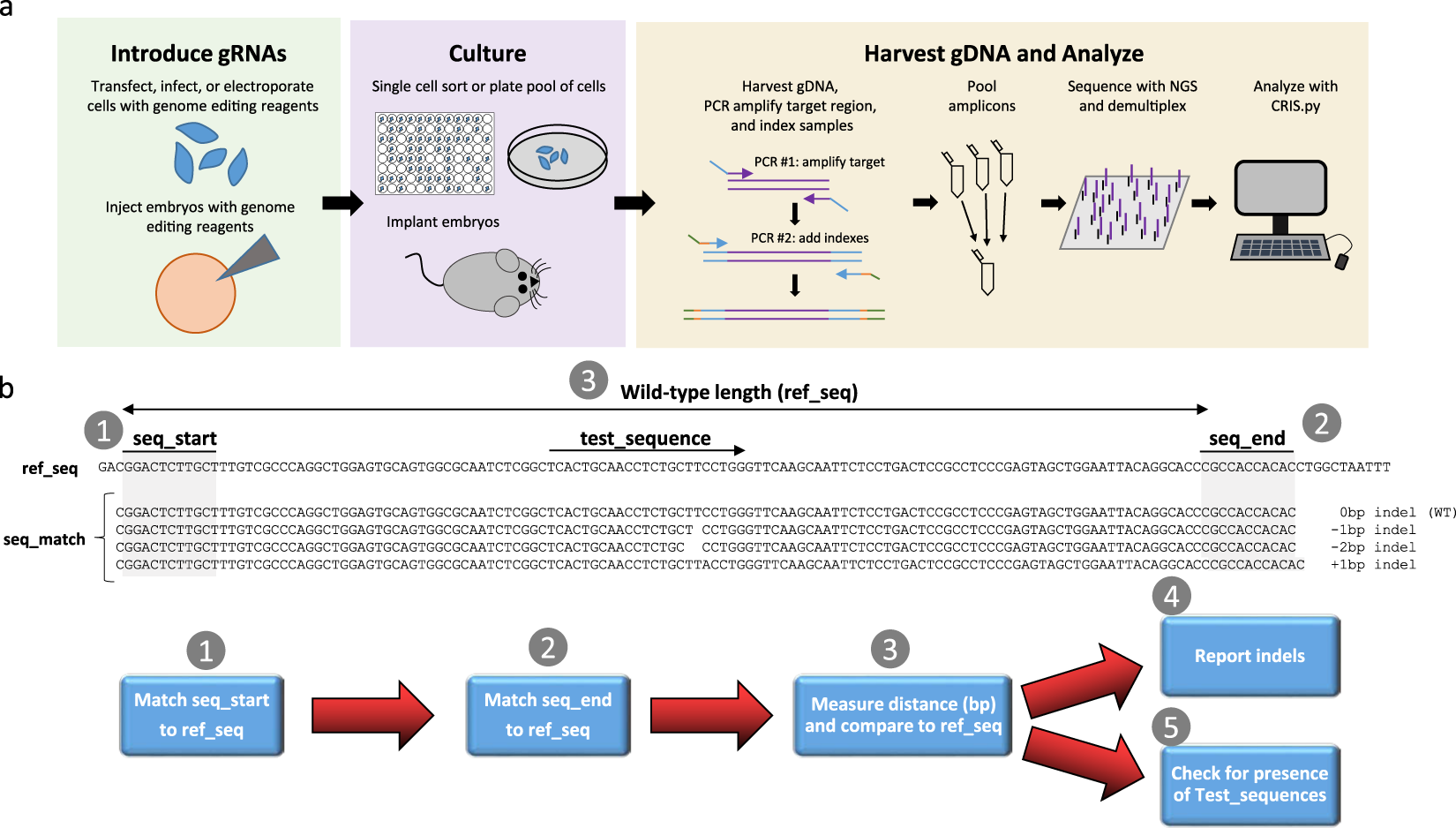
CRIS.py: A Versatile and High-throughput Analysis Program for CRISPR-based Genome Editing | Scientific Reports
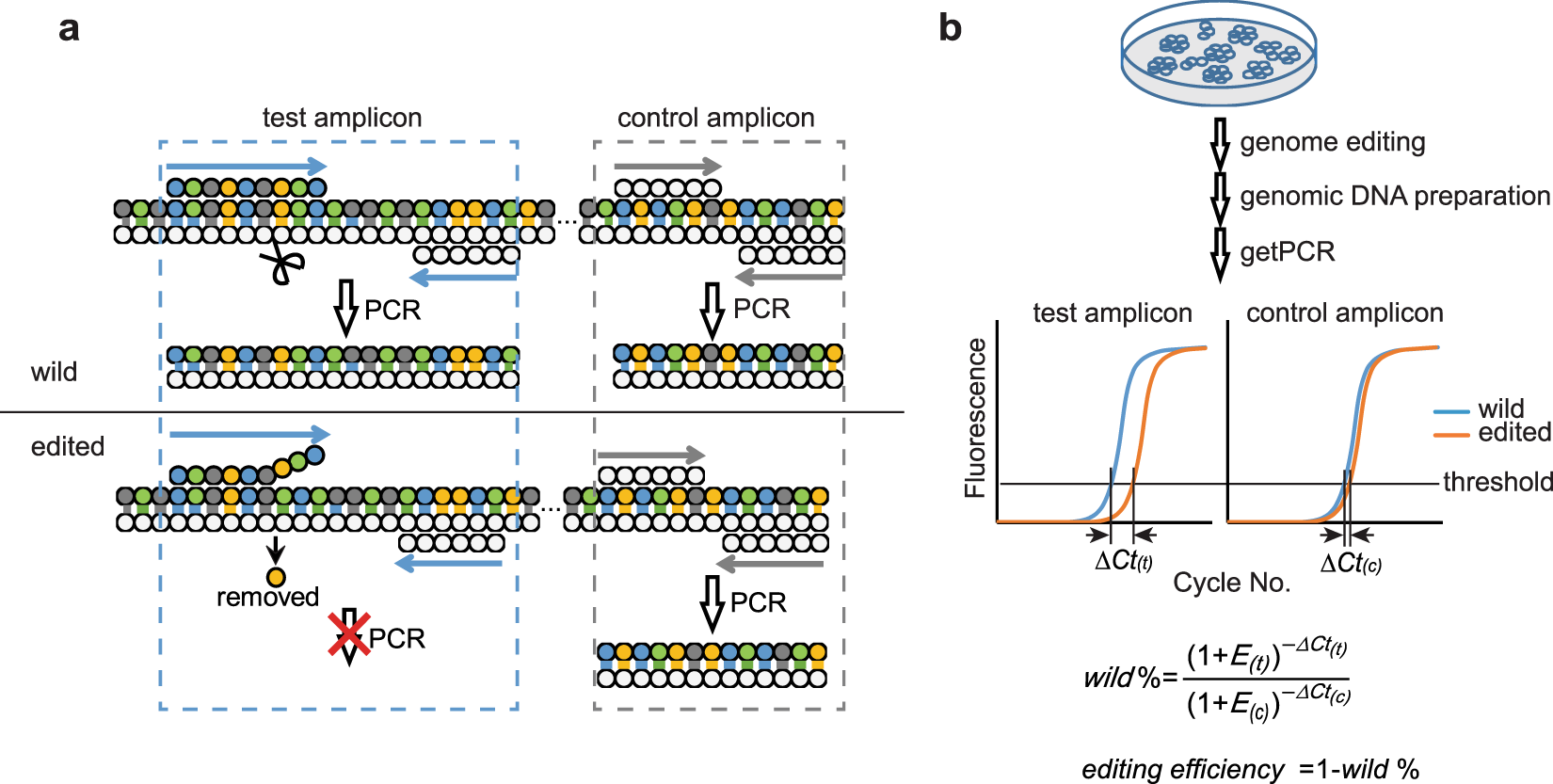
A qPCR method for genome editing efficiency determination and single-cell clone screening in human cells | Scientific Reports