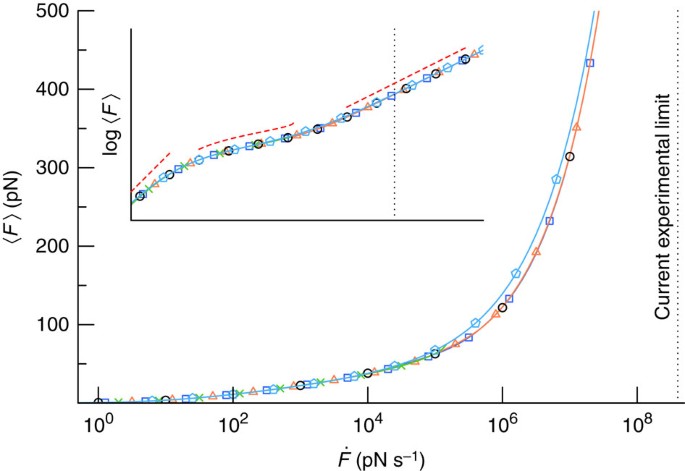
Bell-Evans plot of unfolding and unbinding forces. For all specific... | Download Scientific Diagram

Extending Bell's Model: How Force Transducer Stiffness Alters Measured Unbinding Forces and Kinetics of Molecular Complexes - ScienceDirect
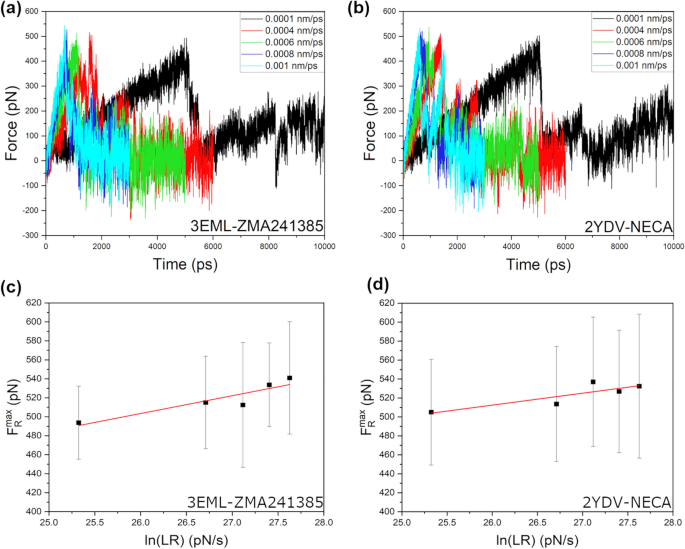
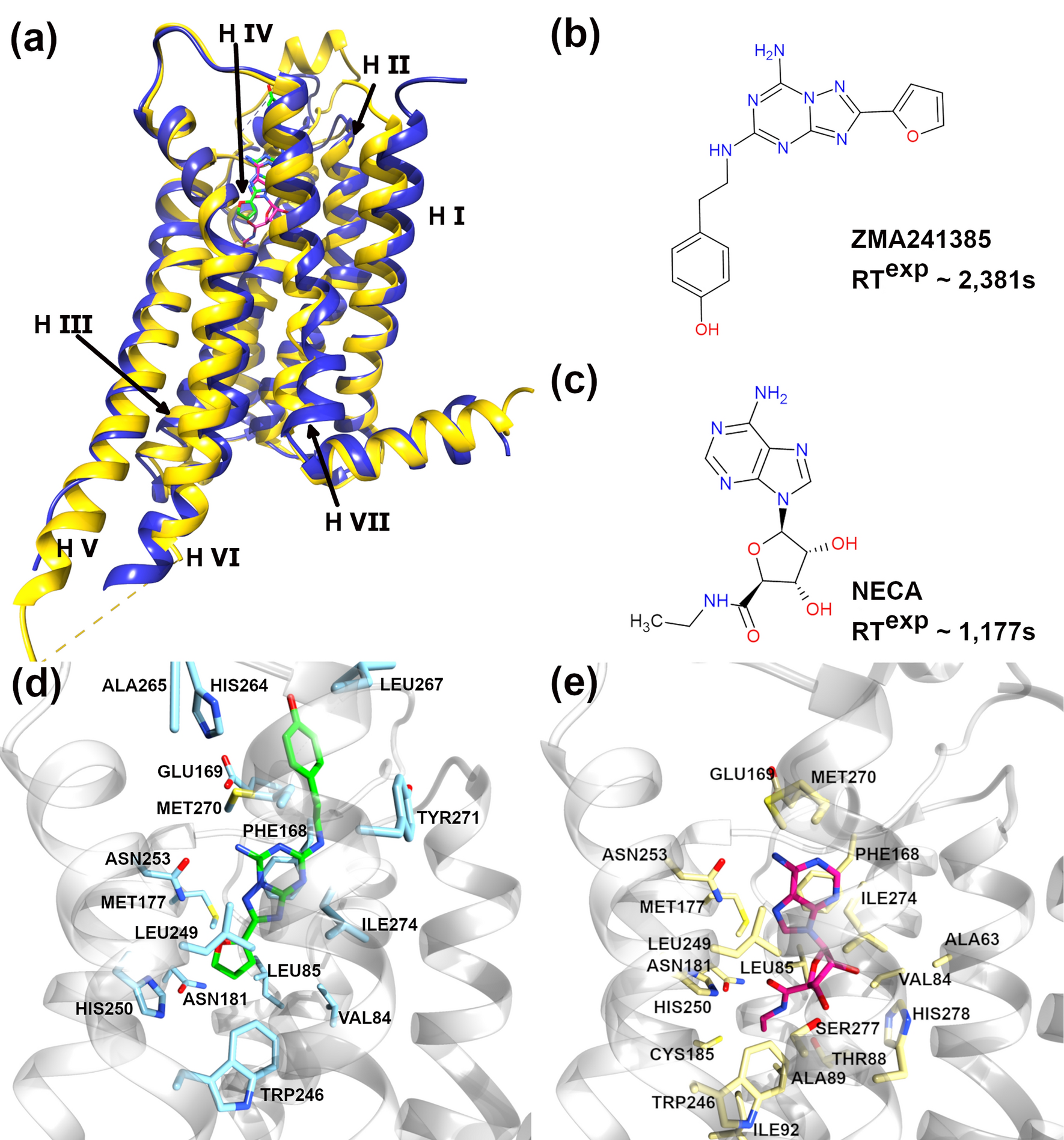
Bell-Evans model and steered molecular dynamics in uncovering the dissociation kinetics of ligands targeting G-protein-coupled receptors | Scientific Reports
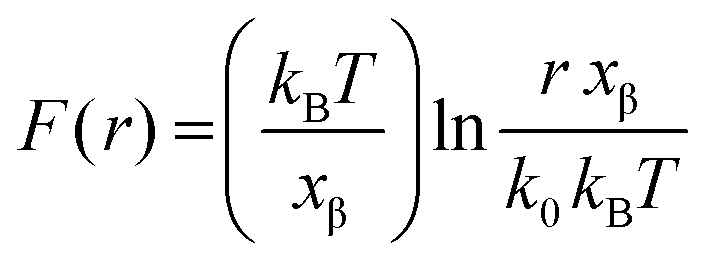
Comparison of three competing dynamic force spectroscopy models to study binding forces of amyloid-β (1–42) - Soft Matter (RSC Publishing) DOI:10.1039/C3SM52257A

The application of atomic force spectroscopy to the study of biological complexes undergoing a biorecognition process. | Semantic Scholar
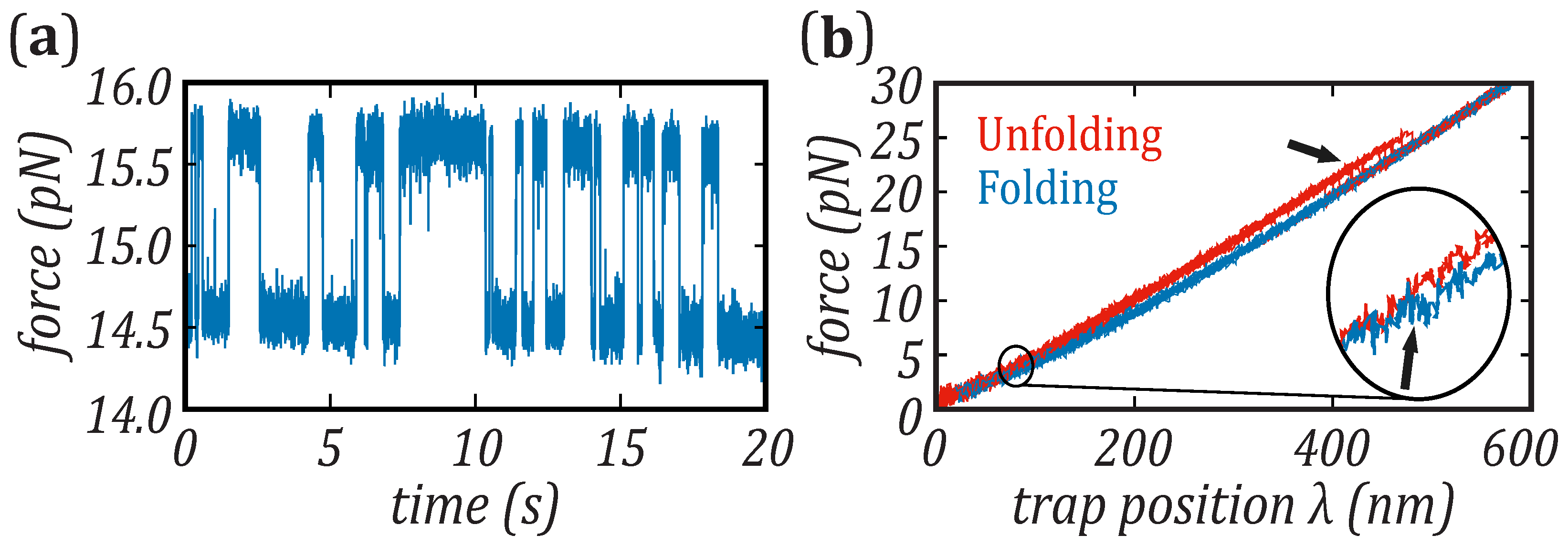
Nanomaterials | Free Full-Text | Force Dependence of Proteins' Transition State Position and the Bell–Evans Model
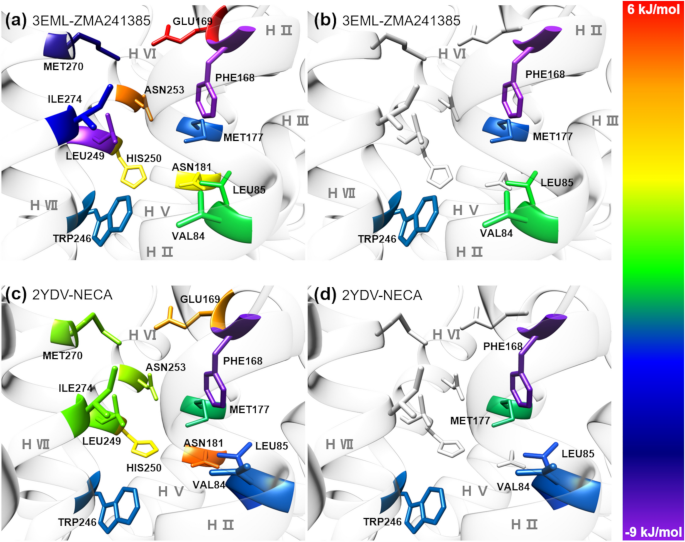
Bell-Evans model and steered molecular dynamics in uncovering the dissociation kinetics of ligands targeting G-protein-coupled receptors | Scientific Reports

Bell-Evans plot showing a linear relationship between the most probable... | Download Scientific Diagram

A) Rupture force vs. loading rate using the Bell – Evans model. Most... | Download Scientific Diagram

Kinetic parameters derived from the Bell-Evans model and applied to the... | Download Scientific Diagram

Nanomaterials | Free Full-Text | Force Dependence of Proteins' Transition State Position and the Bell–Evans Model
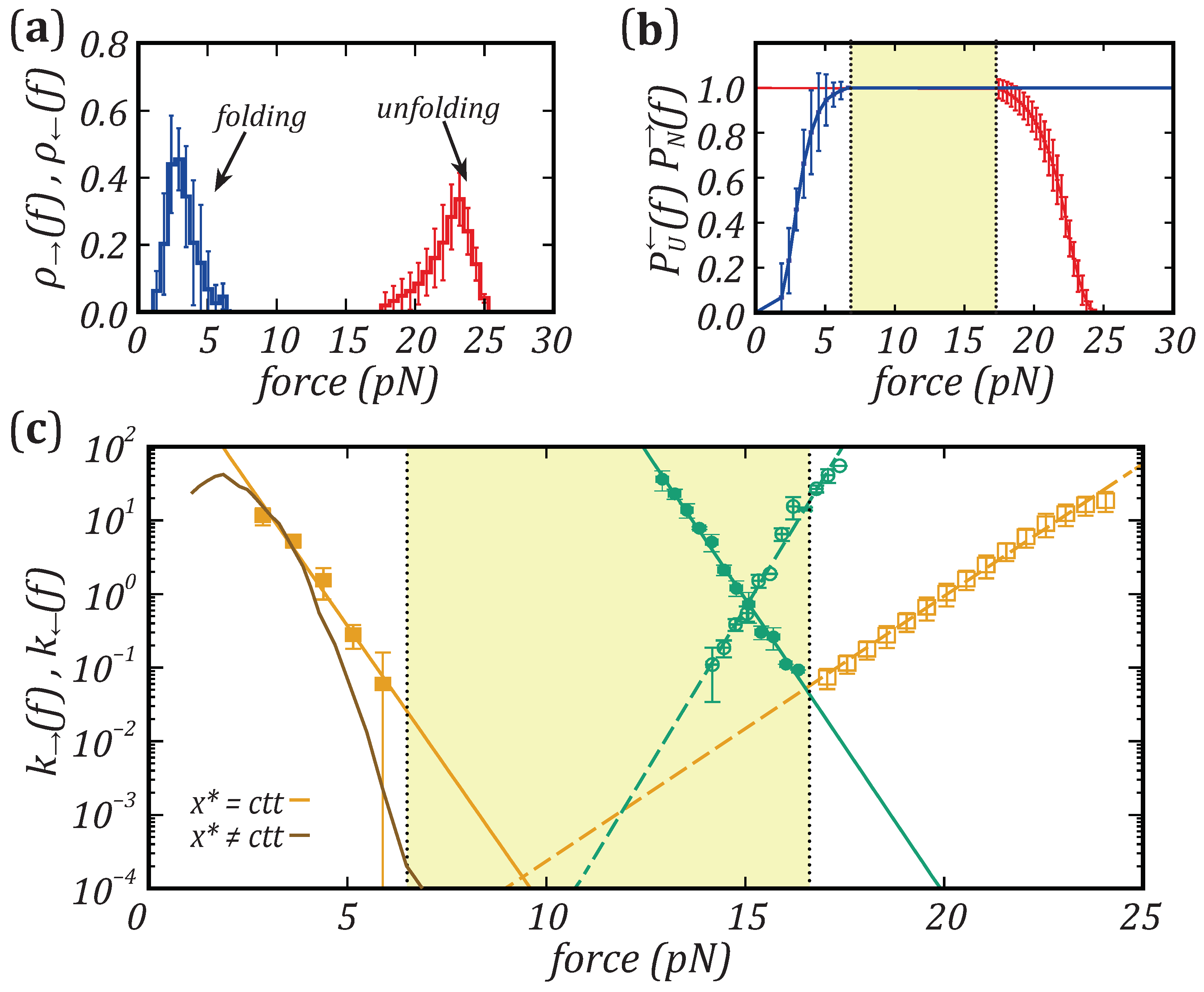
Nanomaterials | Free Full-Text | Force Dependence of Proteins' Transition State Position and the Bell–Evans Model
Effects of Multiple-Bond Ruptures on Kinetic Parameters Extracted from Force Spectroscopy Measurements: Revisiting Biotin-Strept
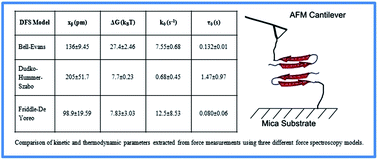
Comparison of three competing dynamic force spectroscopy models to study binding forces of amyloid-β (1–42) - Soft Matter (RSC Publishing)
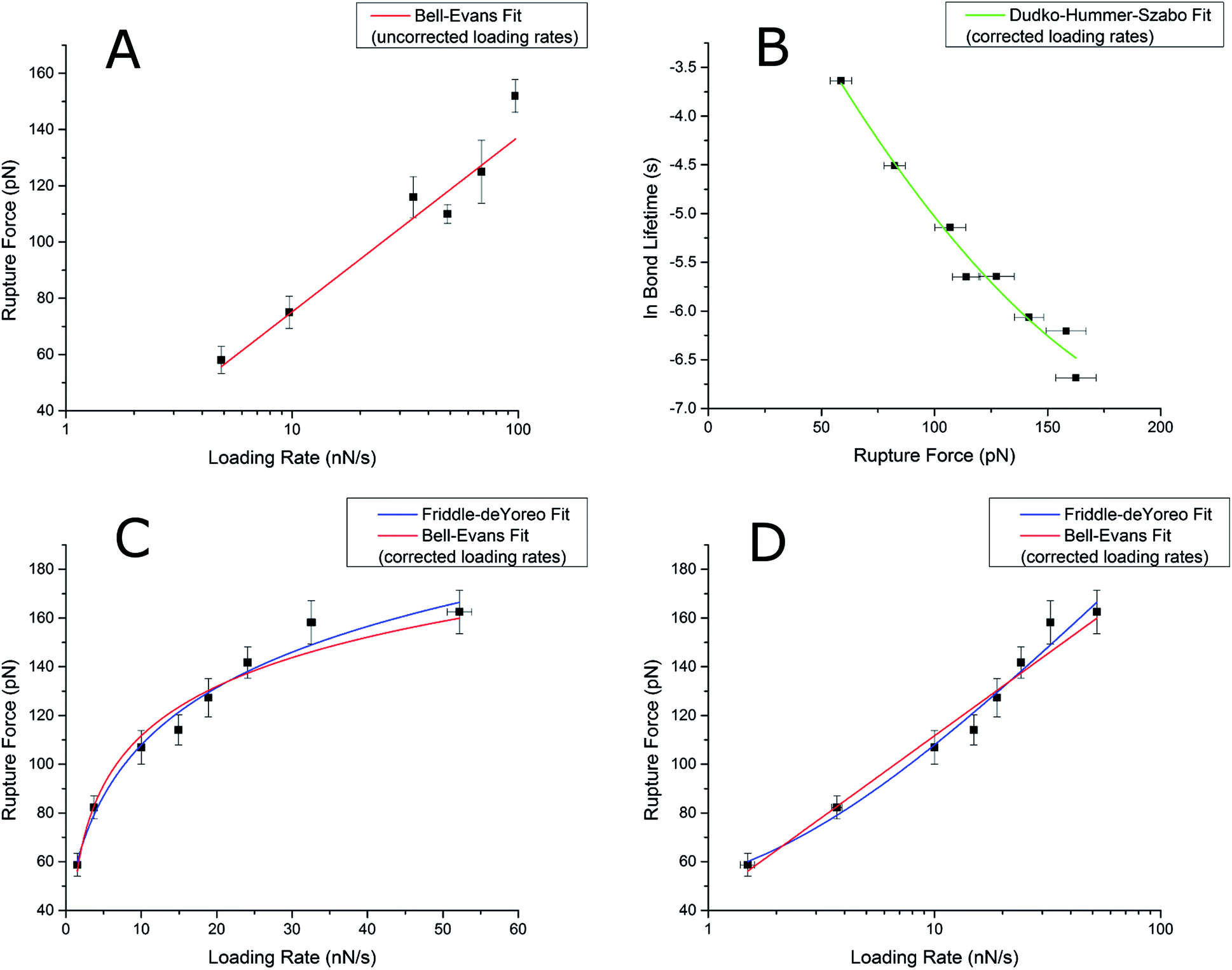
Comparison of three competing dynamic force spectroscopy models to study binding forces of amyloid-β (1–42) - Soft Matter (RSC Publishing) DOI:10.1039/C3SM52257A

Figure 5 from Bell-Evans-Polanyi principle for molecular dynamics trajectories and its implications for global optimization. | Semantic Scholar
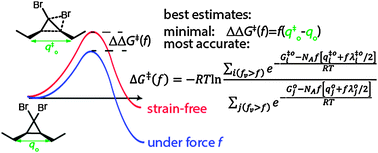
Comparison of the predictive performance of the Bell–Evans, Taylor-expansion and statistical-mechanics models of mechanochemistry - Chemical Communications (RSC Publishing)