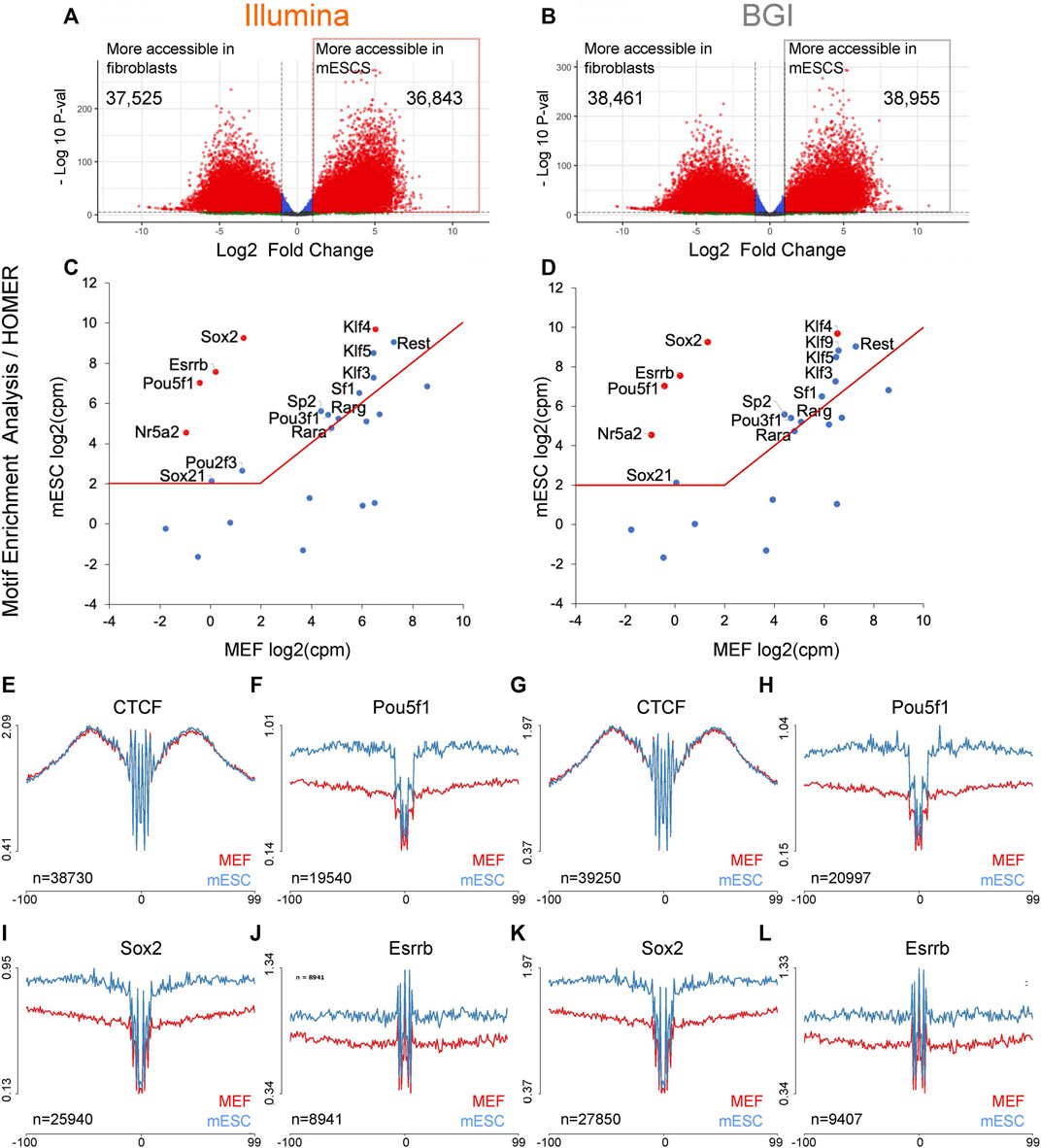
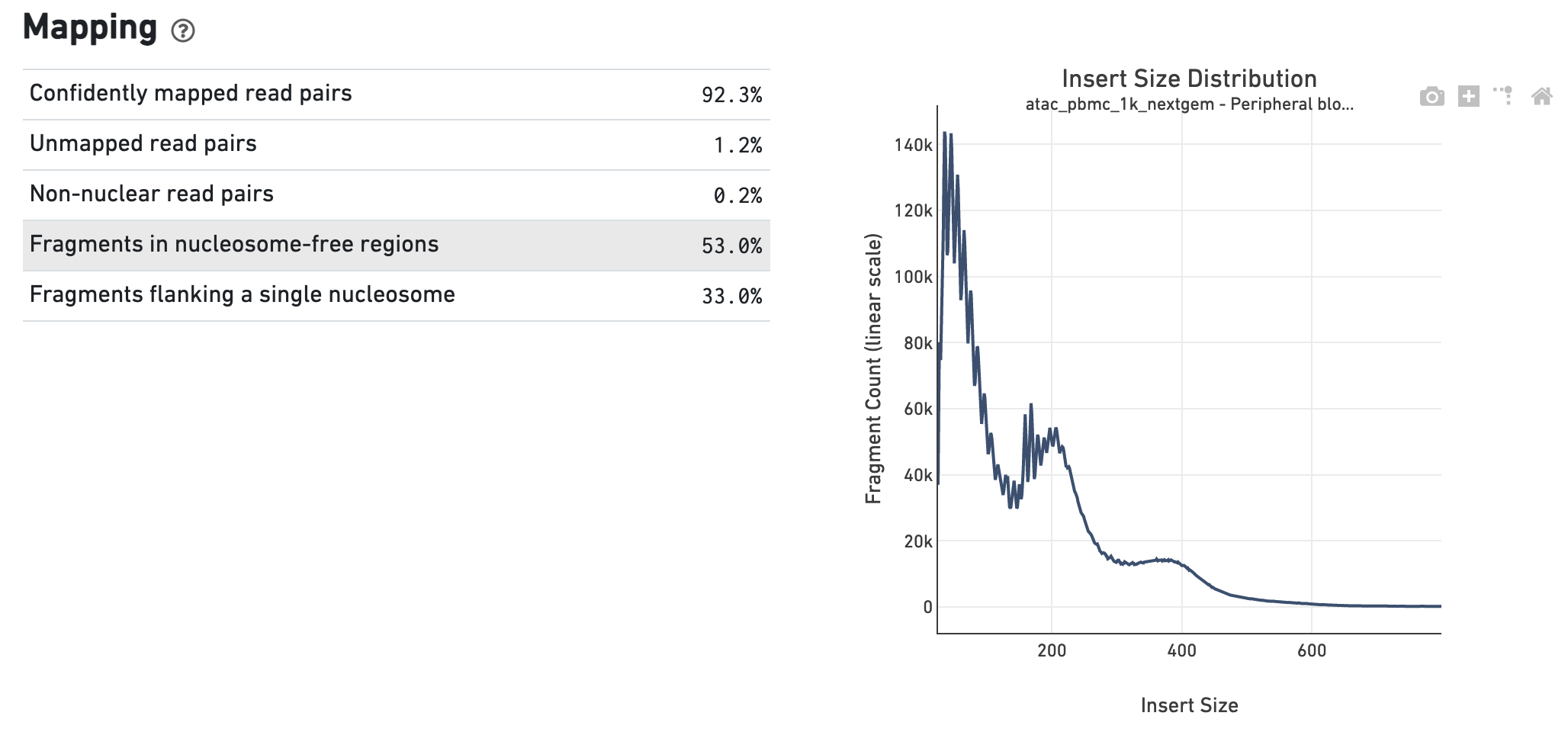
Quantification, Dynamic Visualization, and Validation of Bias in ATAC-Seq Data with ataqv - ScienceDirect

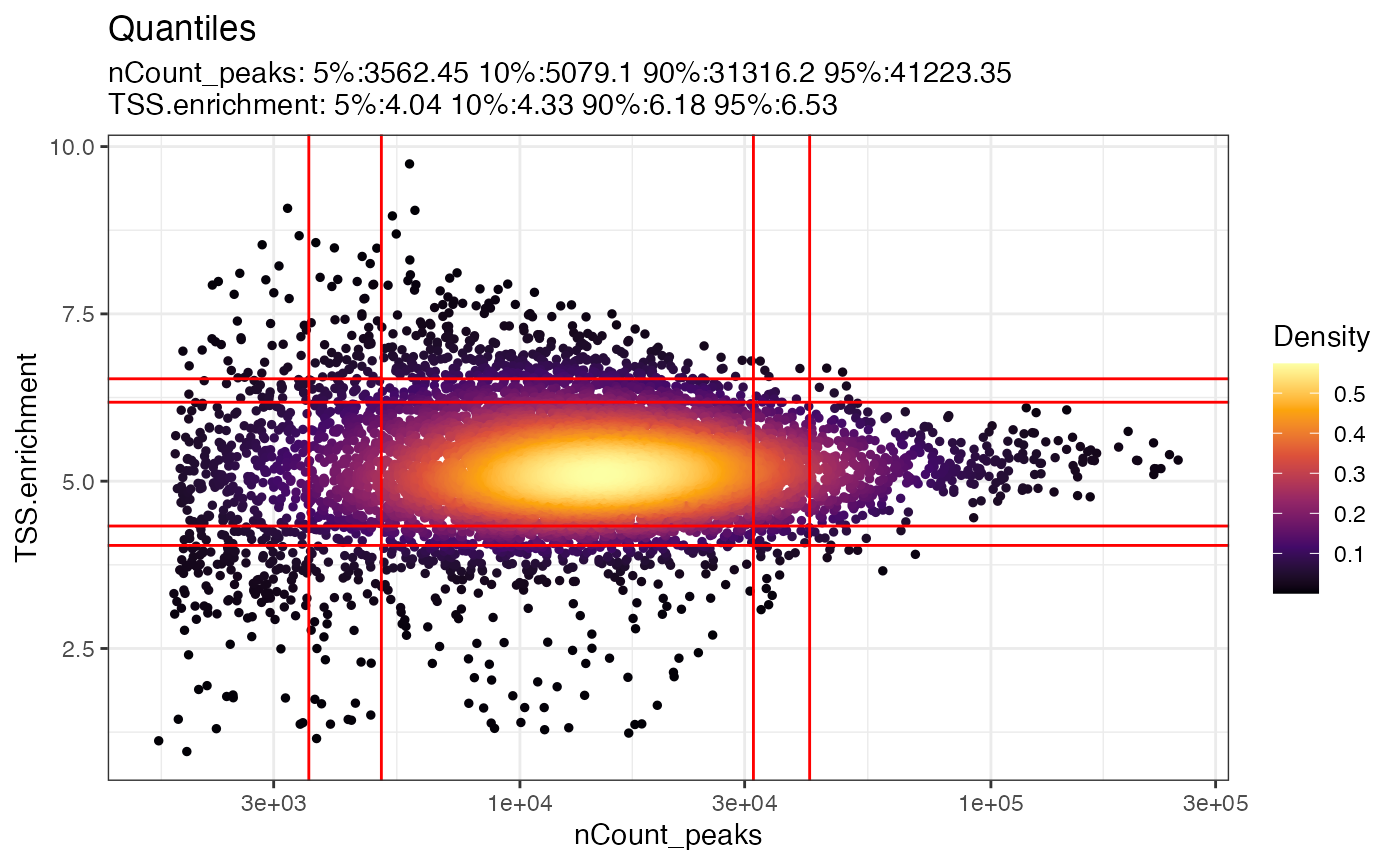
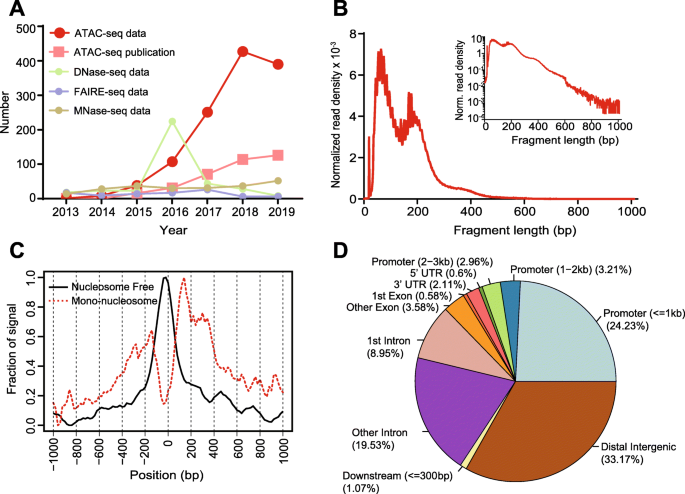
ATAC-seq data quality control and characteristics. (A) Distribution of... | Download Scientific Diagram
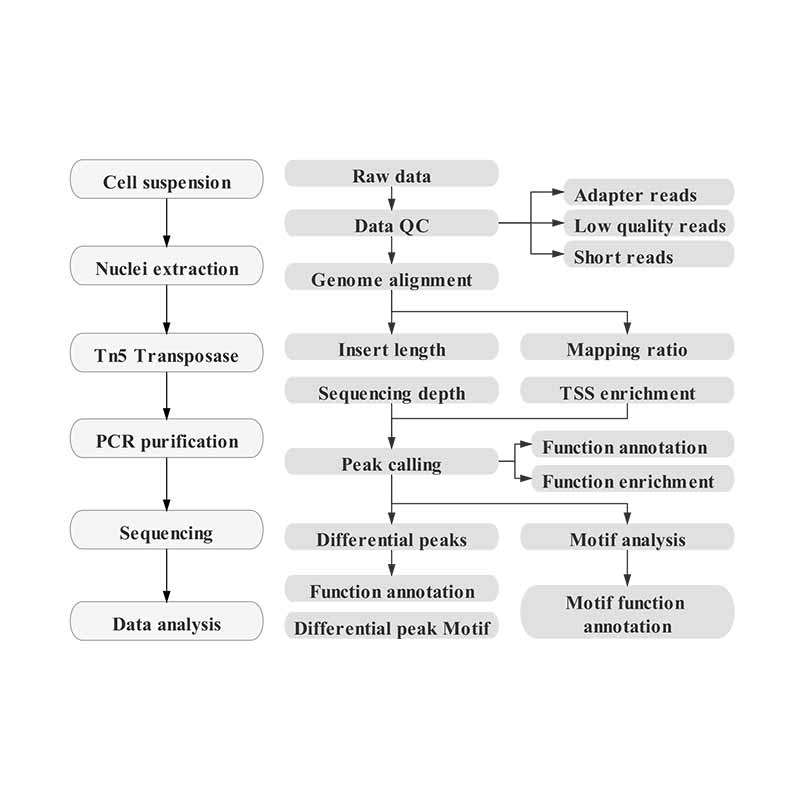
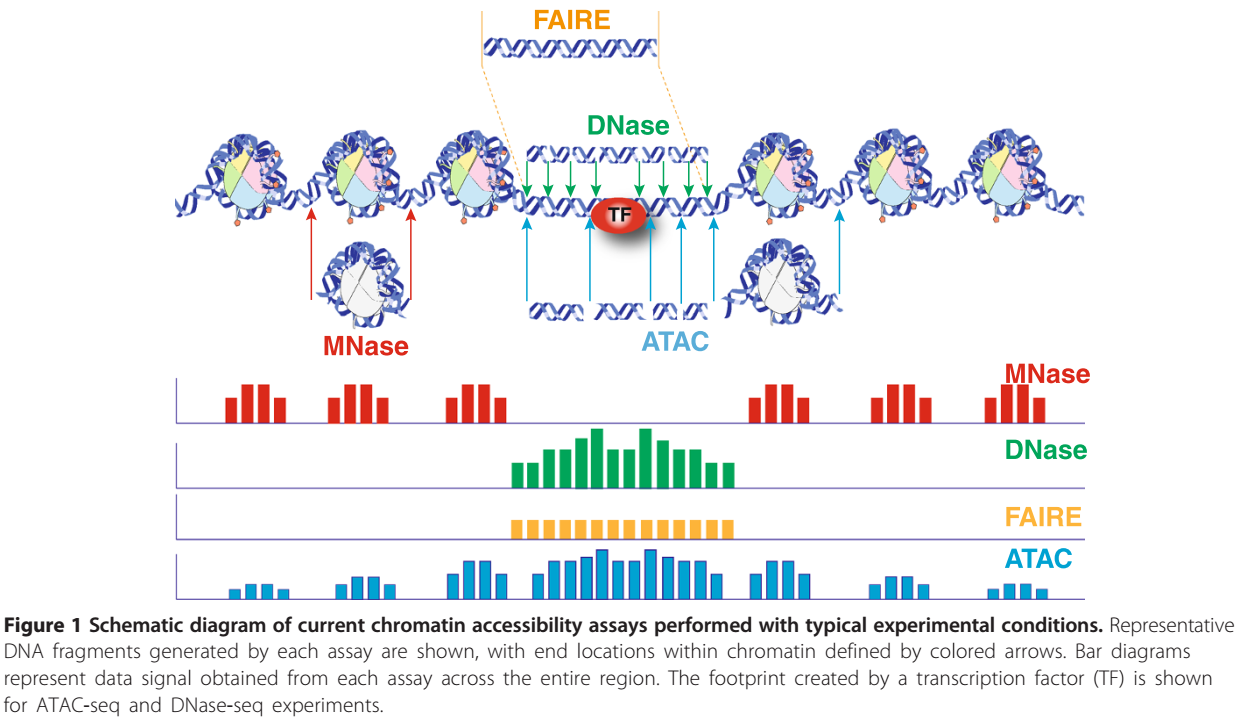
China Assay für Transposase-zugängliches Chromatin mit Hochdurchsatz-Sequenzierung (ATAC-seq) Fabrik und Hersteller |BMK
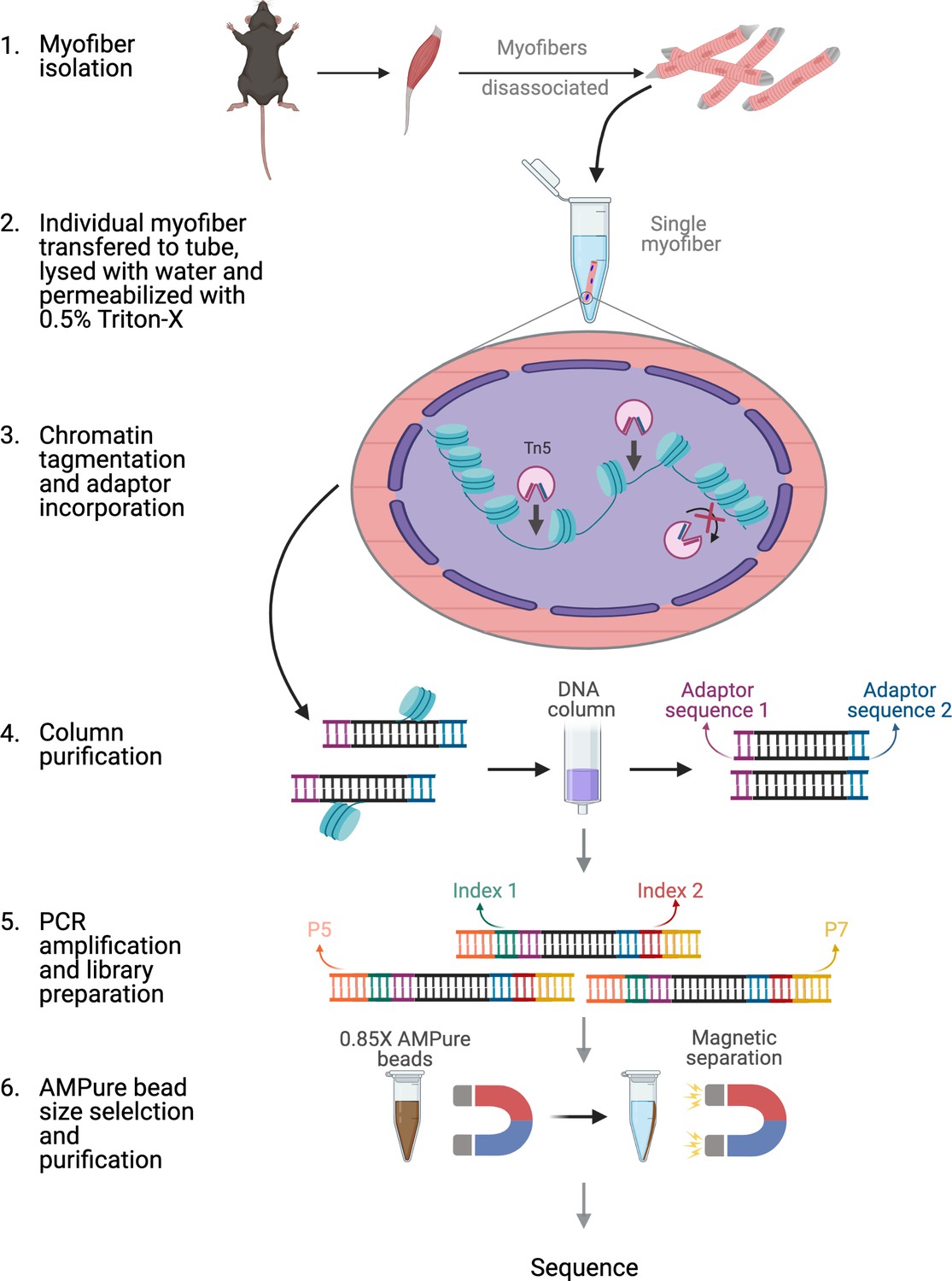
Application of ATAC-Seq for genome-wide analysis of the chromatin state at single myofiber resolution | eLife
Clustering and identification of cell-type clusters in sci-ATAC-seq... | Download Scientific Diagram
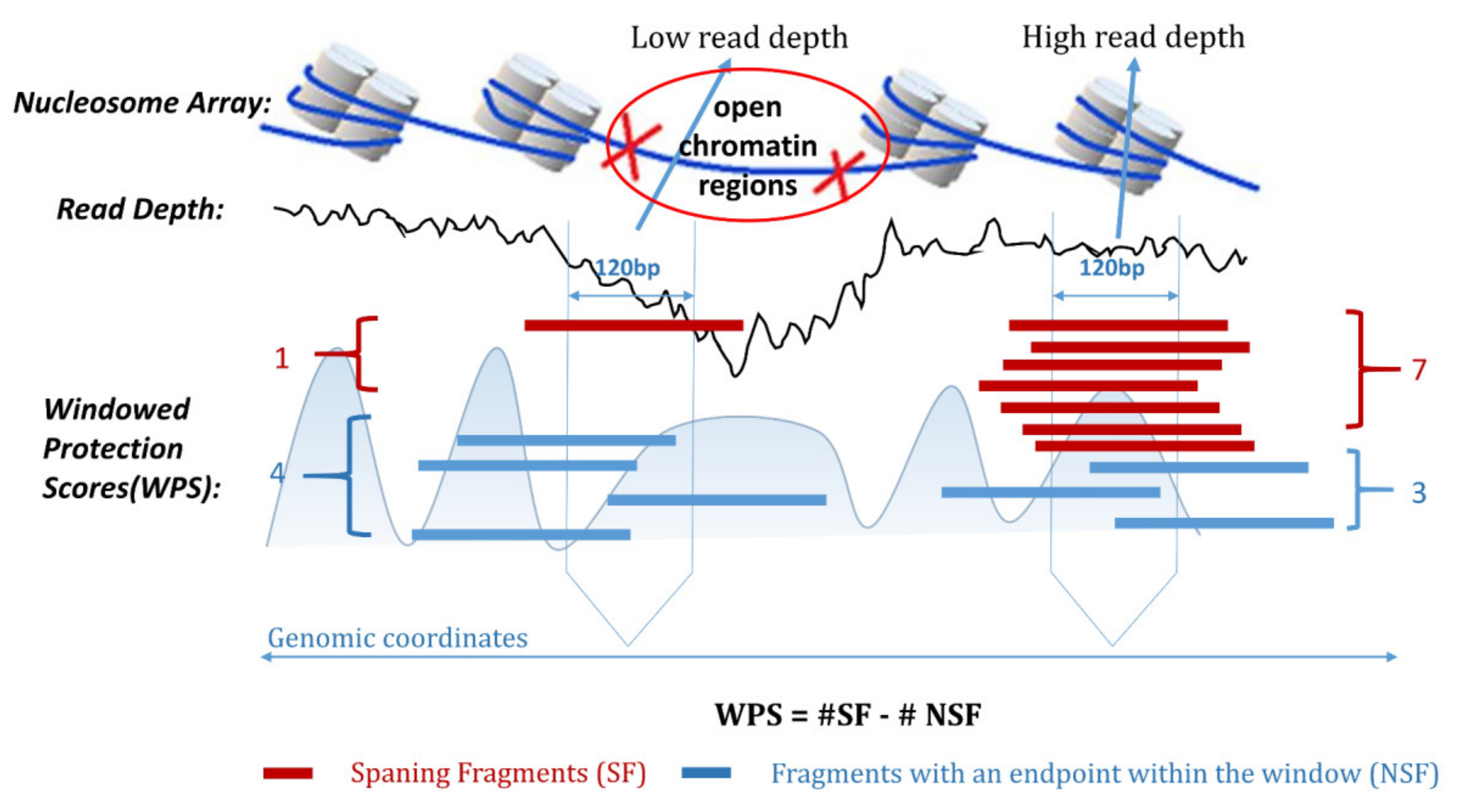
IJMS | Free Full-Text | OCRDetector: Accurately Detecting Open Chromatin Regions via Plasma Cell-Free DNA Sequencing Data

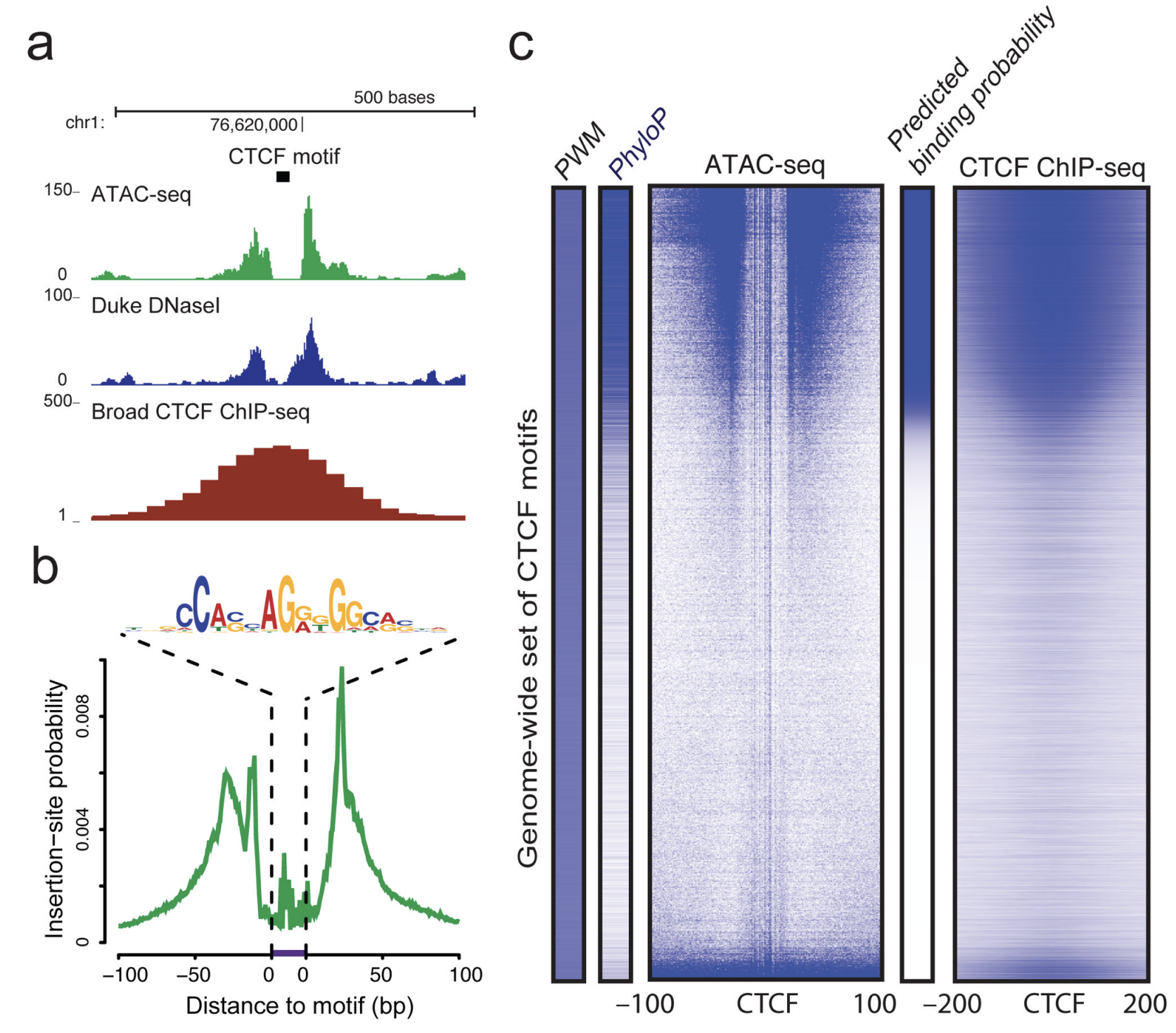
ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation | Nature Communications

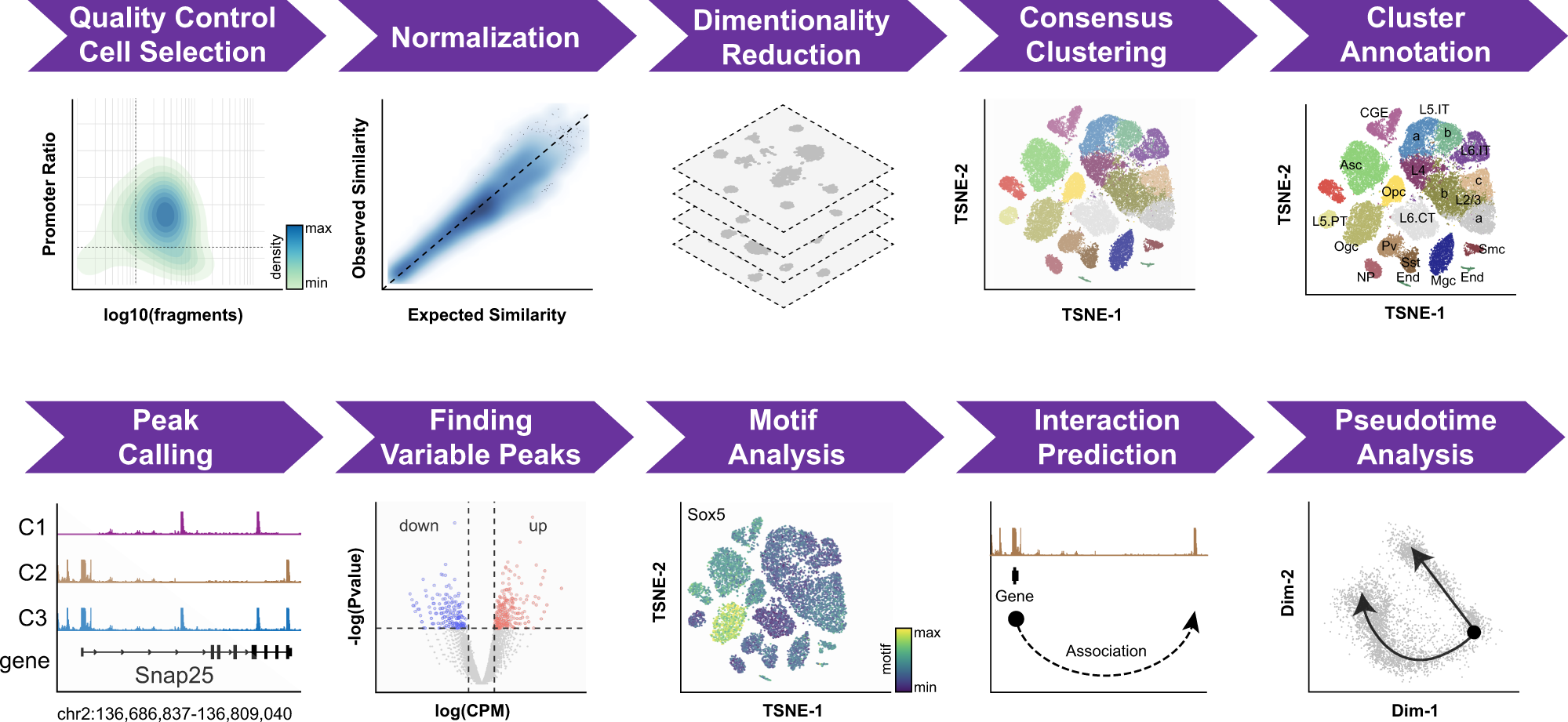
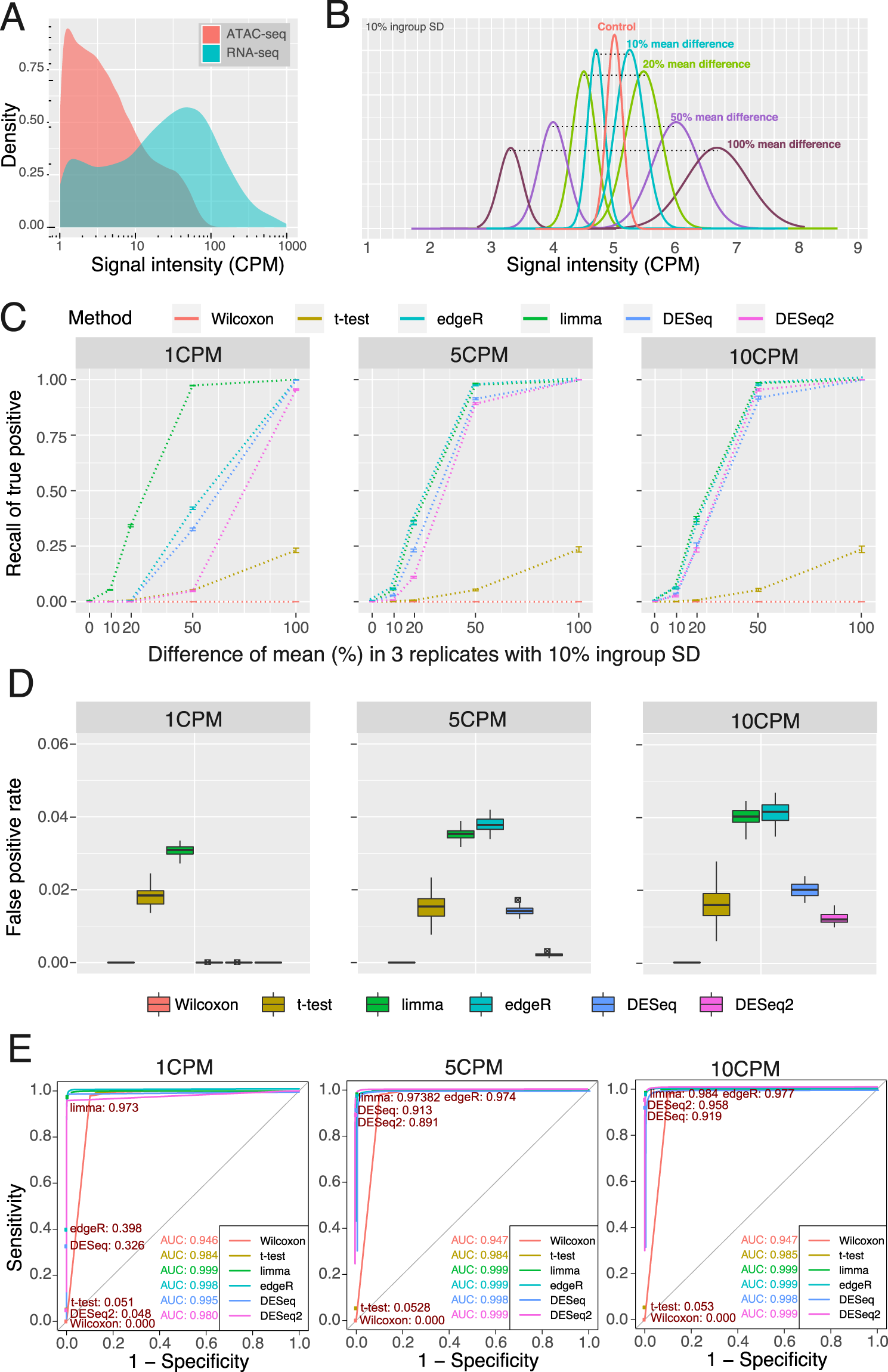
Single-cell ATAC sequencing analysis: From data preprocessing to hypothesis generation - ScienceDirect

ATAC-seq data QC a, Peak calling versus library sequencing depth. We... | Download Scientific Diagram
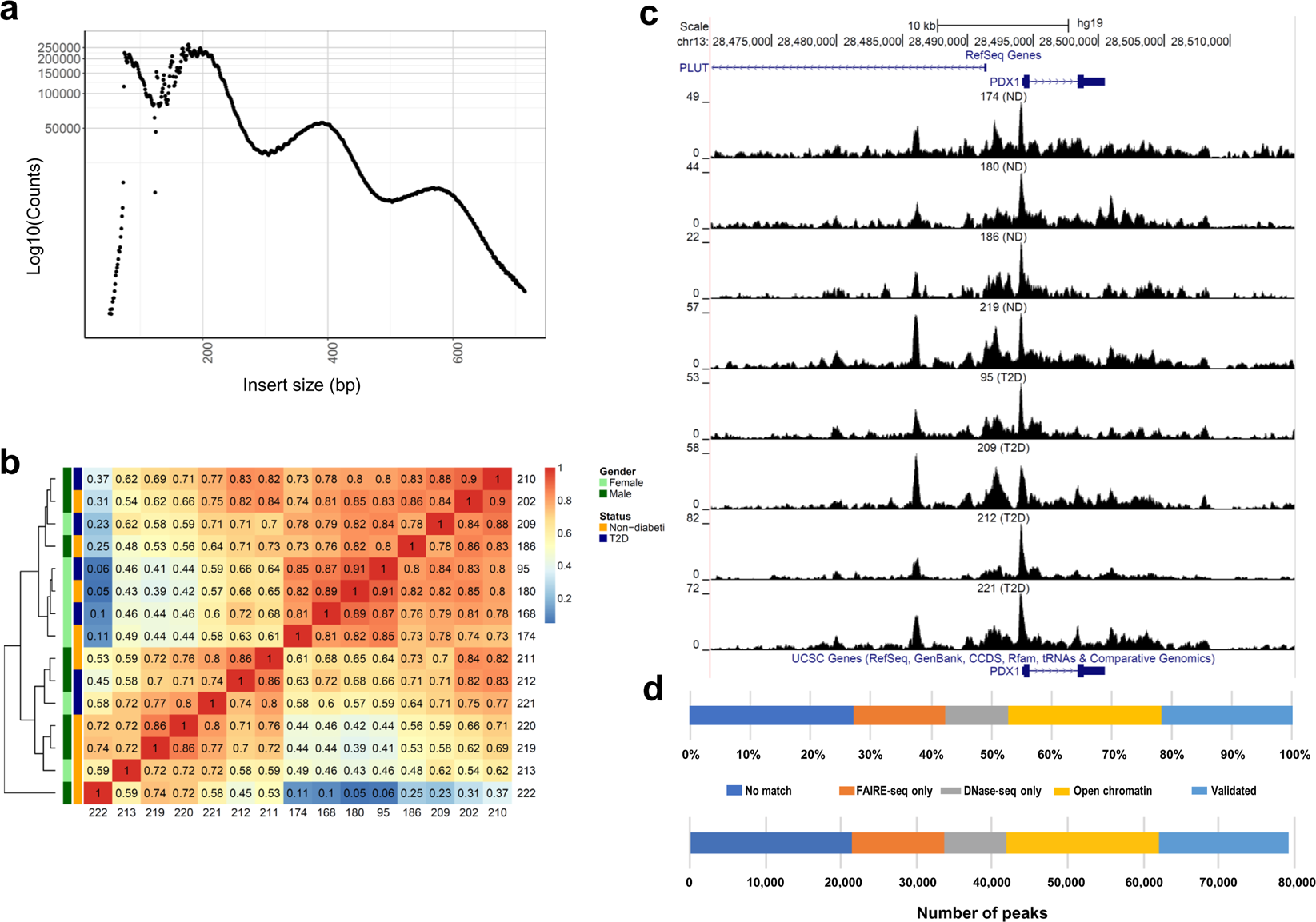
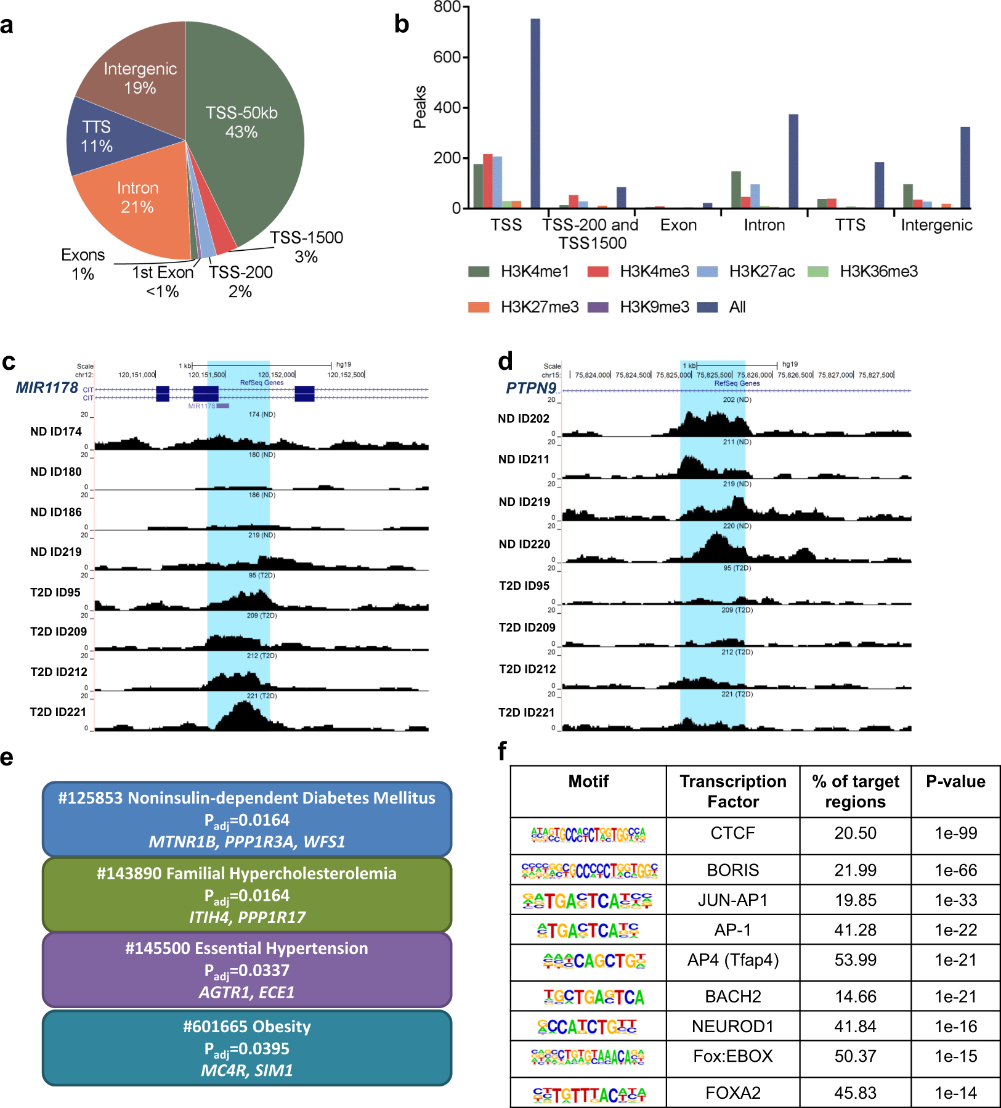
ATAC-seq reveals alterations in open chromatin in pancreatic islets from subjects with type 2 diabetes | Scientific Reports

An optimized ATAC-seq protocol for genome-wide mapping of active regulatory elements in primary mouse cortical neurons